A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting
- PMID: 27461529
- PMCID: PMC4974459
- DOI: 10.1038/ncomms12248
A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting
Abstract
Live-cell imaging has revealed unexpected features of gene expression. Here using improved single-molecule RNA microscopy, we show that synthesis of HIV-1 RNA is achieved by groups of closely spaced polymerases, termed convoys, as opposed to single isolated enzymes. Convoys arise by a Mediator-dependent reinitiation mechanism, which generates a transient but rapid succession of polymerases initiating and escaping the promoter. During elongation, polymerases are spaced by few hundred nucleotides, and physical modelling suggests that DNA torsional stress may maintain polymerase spacing. We additionally observe that the HIV-1 promoter displays stochastic fluctuations on two time scales, which we refer to as multi-scale bursting. Each time scale is regulated independently: Mediator controls minute-scale fluctuation (convoys), while TBP-TATA-box interaction controls sub-hour fluctuations (long permissive/non-permissive periods). A cellular promoter also produces polymerase convoys and displays multi-scale bursting. We propose that slow, TBP-dependent fluctuations are important for phenotypic variability of single cells.
Figures
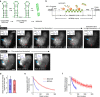
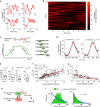
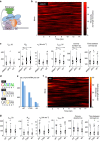



Comment in
-
Gene Regulation: Activation through Space.Curr Biol. 2016 Oct 10;26(19):R895-R898. doi: 10.1016/j.cub.2016.08.031. Epub 2016 Oct 10. Curr Biol. 2016. PMID: 27728794
Similar articles
-
Evidence for functional interaction between the HIV-1 Tat transactivator and the TATA box binding protein in vivo.J Mol Biol. 1995 Jul 7;250(2):169-80. doi: 10.1006/jmbi.1995.0368. J Mol Biol. 1995. PMID: 7608968
-
Distinct transcriptional responses of RNA polymerases I, II and III to aptamers that bind TBP.Nucleic Acids Res. 2005 Feb 8;33(3):838-45. doi: 10.1093/nar/gki212. Print 2005. Nucleic Acids Res. 2005. PMID: 15701755 Free PMC article.
-
HIV-1 Tat stimulates transcription complex assembly through recruitment of TBP in the absence of TAFs.PLoS Biol. 2005 Feb;3(2):e44. doi: 10.1371/journal.pbio.0030044. Epub 2005 Feb 8. PLoS Biol. 2005. PMID: 15719058 Free PMC article.
-
The Role of Supercoiling in the Motor Activity of RNA Polymerases.Methods Mol Biol. 2018;1805:215-232. doi: 10.1007/978-1-4939-8556-2_11. Methods Mol Biol. 2018. PMID: 29971720 Review.
-
Probing steps in DNA transcription using single-molecule methods.J Biol Chem. 2021 Sep;297(3):101086. doi: 10.1016/j.jbc.2021.101086. Epub 2021 Aug 14. J Biol Chem. 2021. PMID: 34403697 Free PMC article. Review.
Cited by
-
The Stochastic Genome and Its Role in Gene Expression.Cold Spring Harb Perspect Biol. 2021 Oct 1;13(10):a040386. doi: 10.1101/cshperspect.a040386. Cold Spring Harb Perspect Biol. 2021. PMID: 33903158 Free PMC article. Review.
-
Live-cell imaging reveals the spatiotemporal organization of endogenous RNA polymerase II phosphorylation at a single gene.Nat Commun. 2021 May 26;12(1):3158. doi: 10.1038/s41467-021-23417-0. Nat Commun. 2021. PMID: 34039974 Free PMC article.
-
Technologies Enabling Single-Molecule Super-Resolution Imaging of mRNA.Cells. 2022 Sep 30;11(19):3079. doi: 10.3390/cells11193079. Cells. 2022. PMID: 36231040 Free PMC article. Review.
-
Nascent Transcript Folding Plays a Major Role in Determining RNA Polymerase Elongation Rates.Mol Cell. 2020 Aug 6;79(3):488-503.e11. doi: 10.1016/j.molcel.2020.06.002. Epub 2020 Jun 24. Mol Cell. 2020. PMID: 32585128 Free PMC article.
-
A Stochastic Model of Gene Expression with Polymerase Recruitment and Pause Release.Biophys J. 2020 Sep 1;119(5):1002-1014. doi: 10.1016/j.bpj.2020.07.020. Epub 2020 Aug 3. Biophys J. 2020. PMID: 32814062 Free PMC article.
References
-
- Kandiah E., Trowitzsch S., Gupta K., Haffke M. & Berger I. More pieces to the puzzle: recent structural insights into class II transcription initiation. Curr. Opin. Struct. Biol. 24, 91–97 (2014). - PubMed
-
- Thomas M. & Chiang C. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41, 105–178 (2006). - PubMed
-
- Dynlacht B., Hoey T. & Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell 66, 563–576 (1991). - PubMed
-
- Pugh B. Control of gene expression through regulation of the TATA-binding protein. Gene 255, 1–14 (2000). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

