Targeting cell membrane adaptation as a novel antimicrobial strategy
- PMID: 27458841
- PMCID: PMC5119629
- DOI: 10.1016/j.mib.2016.07.002
Targeting cell membrane adaptation as a novel antimicrobial strategy
Abstract
Emergence of antibiotic resistance is an example of the incredible plasticity of bacteria to survive in all environments. The search for new antibiotics active against traditional targets is more challenging due not only to the lack of novel natural products to fulfill the current clinical needs against multidrug-resistant (MDR) bacteria, but also for the possible 'collateral' effects on the human microbiota. Thus, non-traditional approaches to combat MDR bacteria have been proposed. Here, we discuss the possibility of targeting the membrane response to the antibiotic attack (cell membrane adaptation) as a viable strategy to increase the activity of current antimicrobials, enhance the activity of the innate immune system and prevent development of resistance during therapy using the three-component regulatory system LiaFSR of enterococci as a model.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures
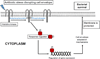
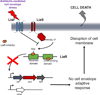
Similar articles
-
A liaR deletion restores susceptibility to daptomycin and antimicrobial peptides in multidrug-resistant Enterococcus faecalis.J Infect Dis. 2015 Apr 15;211(8):1317-25. doi: 10.1093/infdis/jiu602. Epub 2014 Oct 31. J Infect Dis. 2015. PMID: 25362197 Free PMC article.
-
Evaluation of membrane fluidity of multidrug-resistant isolates of Escherichia coli and Staphylococcus aureus in presence and absence of antibiotics.J Photochem Photobiol B. 2018 Apr;181:150-156. doi: 10.1016/j.jphotobiol.2018.03.002. Epub 2018 Mar 5. J Photochem Photobiol B. 2018. PMID: 29567316
-
Polyacylated neohesperidosides from Geranium caespitosum: bacterial multidrug resistance pump inhibitors.Bioorg Med Chem Lett. 2003 Jun 2;13(11):1915-8. doi: 10.1016/s0960-894x(03)00316-0. Bioorg Med Chem Lett. 2003. PMID: 12749897
-
Multidrug-Resistant Enterococcal Infections: New Compounds, Novel Antimicrobial Therapies?Trends Microbiol. 2017 Jun;25(6):467-479. doi: 10.1016/j.tim.2017.01.004. Epub 2017 Feb 13. Trends Microbiol. 2017. PMID: 28209400 Review.
-
Plant essential oils and their constituents in coping with multidrug-resistant bacteria.Expert Rev Anti Infect Ther. 2012 Jul;10(7):775-90. doi: 10.1586/eri.12.57. Expert Rev Anti Infect Ther. 2012. PMID: 22943401 Review.
Cited by
-
Identification of a Novel Two-Peptide Lantibiotic from Vagococcus fluvialis.Microbiol Spectr. 2022 Aug 31;10(4):e0095422. doi: 10.1128/spectrum.00954-22. Epub 2022 Jun 22. Microbiol Spectr. 2022. PMID: 35730941 Free PMC article.
-
The LiaFSR and BsrXRS Systems Contribute to Bile Salt Resistance in Enterococcus faecium Isolates.Front Microbiol. 2019 May 10;10:1048. doi: 10.3389/fmicb.2019.01048. eCollection 2019. Front Microbiol. 2019. PMID: 31134041 Free PMC article.
-
Human Serum Supplementation Promotes Streptococcus mitis Growth and Induces Specific Transcriptomic Responses.Microbiol Spectr. 2023 Jun 15;11(3):e0512922. doi: 10.1128/spectrum.05129-22. Epub 2023 Apr 4. Microbiol Spectr. 2023. PMID: 37014220 Free PMC article.
-
Mini Review: Bacterial Membrane Composition and Its Modulation in Response to Stress.Front Mol Biosci. 2021 May 11;8:634438. doi: 10.3389/fmolb.2021.634438. eCollection 2021. Front Mol Biosci. 2021. PMID: 34046426 Free PMC article. Review.
-
Two Mutations Commonly Associated with Daptomycin Resistance in Enterococcus faecium LiaST120A and LiaRW73C Appear To Function Epistatically in LiaFSR Signaling.Biochemistry. 2018 Dec 11;57(49):6797-6805. doi: 10.1021/acs.biochem.8b01072. Epub 2018 Nov 27. Biochemistry. 2018. PMID: 30403130 Free PMC article.
References
-
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. - PubMed
-
- Bassetti M, Righi E. Development of novel antibacterial drugs to combat multiple resistant organisms. Langenbecks Arch Surg. 2015;400:153–165. - PubMed
-
- Padmanabhan R, Mishra AK, Raoult D, Fournier PE. Genomics and metagenomics in medical microbiology. J Microbiol Methods. 2013;95:415–424. - PubMed
-
- Klemm E, Dougan G. Advances in understanding bacterial pathogenesis gained from whole-genome sequencing and phylogenetics. Cell Host Microbe. 2016;19:599–610. - PubMed
-
- Mascher T, Margulis NG, Wang T, Ye RW, Helmann JD. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol Microbiol. 2003;50:1591–1604. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

