The effect of DNA extraction methodology on gut microbiota research applications
- PMID: 27456340
- PMCID: PMC4960752
- DOI: 10.1186/s13104-016-2171-7
The effect of DNA extraction methodology on gut microbiota research applications
Abstract
Background: The effect that traditional and modern DNA extraction methods have on applications to study the role of gut microbiota in health and disease is a topic of current interest. Genomic DNA was extracted from three faecal samples and one probiotic capsule using three popular methods; chaotropic (CHAO) method, phenol/chloroform (PHEC) extraction, proprietary kit (QIAG). The performance of each of these methods on DNA yield and quality, microbiota composition using quantitative PCR, deep sequencing of the 16S rRNA gene, and sequencing analysis pipeline was evaluated.
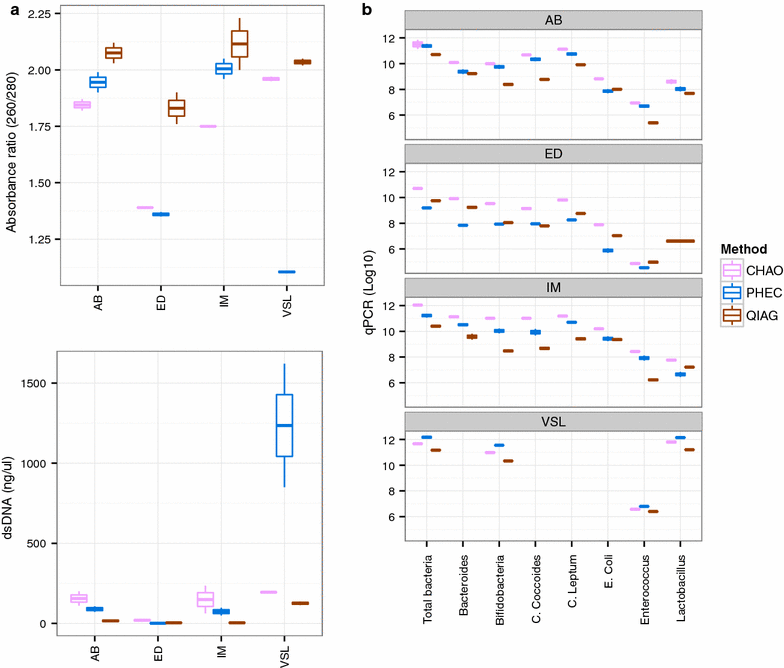
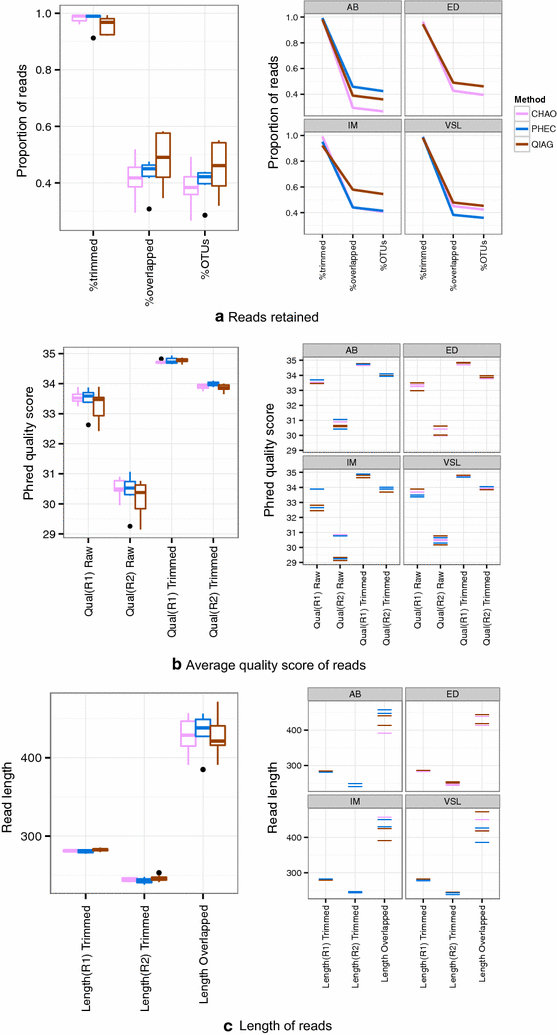
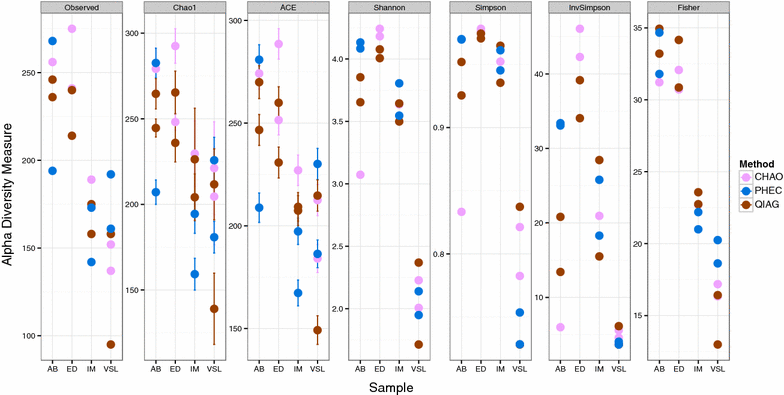
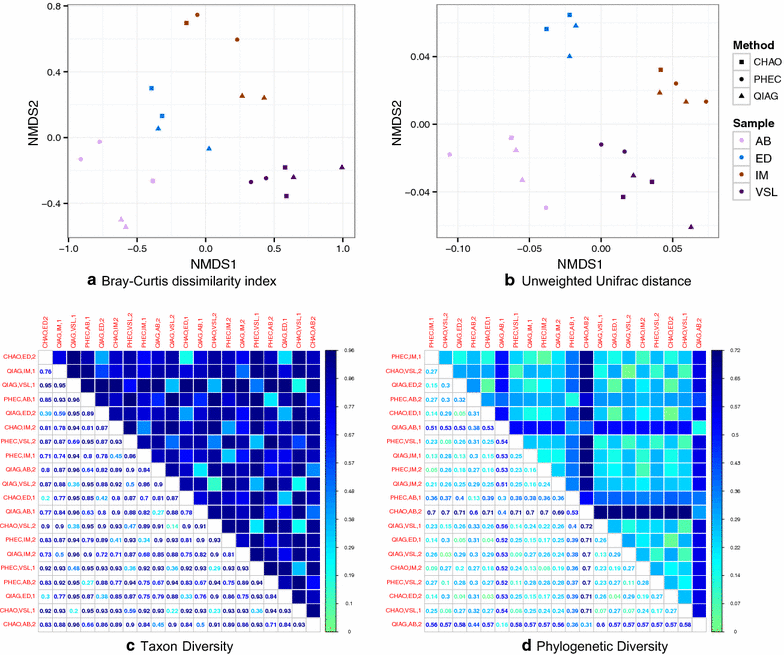
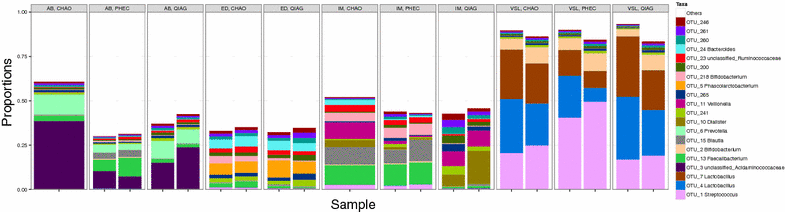
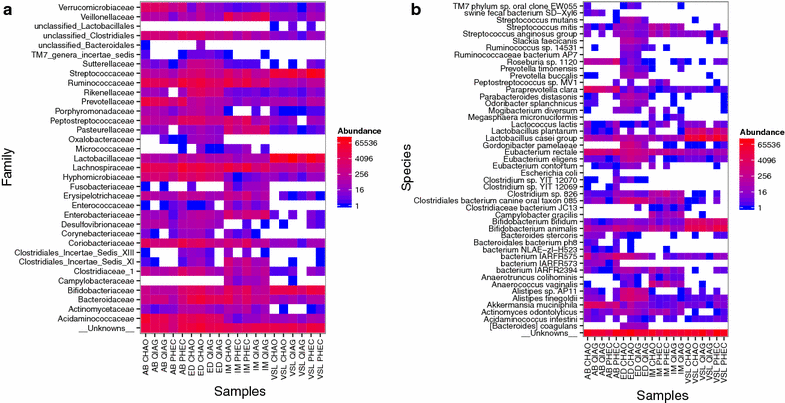
Results: The CHAO yielded the highest and the QIAG kit the lowest amount of double-stranded DNA, but the purity of isolated nucleic acids was better for the latter method. The CHAO method yielded a higher concentration of bacterial taxa per mass (g) of faeces. Sequencing coverage was higher in CHAO method but a higher proportion of the initial sequencing reads were retained for assignments to operational taxonomic unit (OTU) in the QIAG kit compared to the other methods. The QIAG kit appeared to have longer trimmed reads and shorter regions of worse quality than the other two methods. A distinct separation of α-diversity indices between different DNA extraction methods was not observed. When compositional dissimilarities between samples were explored, a strong separation was observed according to sample type. The effect of the extraction method was either marginal (Bray-Curtis distance) or none (unweighted Unifrac distance). Taxon membership and abundance in each sample was independent of the DNA extraction method used.
Conclusions: We have benchmarked several DNA extraction methods commonly used in gut microbiota research and their differences depended on the downstream applications intended for use. Caution should be paid when the intention is to pool and analyse samples or data from studies which have used different DNA extraction methods.
Keywords: 16S rRNA gene; Benchmarking; DNA extraction; Diversity; Metagenomics; PCR.
Figures
Similar articles
-
High-throughput DNA extraction strategy for fecal microbiome studies.Microbiol Spectr. 2024 Jun 4;12(6):e0293223. doi: 10.1128/spectrum.02932-23. Epub 2024 May 15. Microbiol Spectr. 2024. PMID: 38747618 Free PMC article.
-
Impact of DNA extraction, sample dilution, and reagent contamination on 16S rRNA gene sequencing of human feces.Appl Microbiol Biotechnol. 2018 Jan;102(1):403-411. doi: 10.1007/s00253-017-8583-z. Epub 2017 Oct 27. Appl Microbiol Biotechnol. 2018. PMID: 29079861
-
Optimisation of 16S rRNA gut microbiota profiling of extremely low birth weight infants.BMC Genomics. 2017 Nov 2;18(1):841. doi: 10.1186/s12864-017-4229-x. BMC Genomics. 2017. PMID: 29096601 Free PMC article.
-
Evaluation of Methods for the Extraction of Microbial DNA From Vaginal Swabs Used for Microbiome Studies.Front Cell Infect Microbiol. 2019 Jun 6;9:197. doi: 10.3389/fcimb.2019.00197. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31245304 Free PMC article.
-
Metagenomic surveys of gut microbiota.Genomics Proteomics Bioinformatics. 2015 Jun;13(3):148-58. doi: 10.1016/j.gpb.2015.02.005. Epub 2015 Jul 13. Genomics Proteomics Bioinformatics. 2015. PMID: 26184859 Free PMC article. Review.
Cited by
-
Increasing transparency and reproducibility in stroke-microbiota research: A toolbox for microbiota analysis.iScience. 2022 Feb 26;25(4):103998. doi: 10.1016/j.isci.2022.103998. eCollection 2022 Apr 15. iScience. 2022. PMID: 35310944 Free PMC article.
-
Combined bacterial and fungal intestinal microbiota analyses: Impact of storage conditions and DNA extraction protocols.PLoS One. 2018 Aug 3;13(8):e0201174. doi: 10.1371/journal.pone.0201174. eCollection 2018. PLoS One. 2018. PMID: 30074988 Free PMC article.
-
Systematic Bias Introduced by Genomic DNA Template Dilution in 16S rRNA Gene-Targeted Microbiota Profiling in Human Stool Homogenates.mSphere. 2018 Mar 14;3(2):e00560-17. doi: 10.1128/mSphere.00560-17. eCollection 2018 Mar-Apr. mSphere. 2018. PMID: 29564398 Free PMC article.
-
Variation in Rumen Bacteria of Lacaune Dairy Ewes From One Week to the Next.Front Microbiol. 2022 Jun 23;13:848518. doi: 10.3389/fmicb.2022.848518. eCollection 2022. Front Microbiol. 2022. PMID: 35814674 Free PMC article.
-
The Madness of Microbiome: Attempting To Find Consensus "Best Practice" for 16S Microbiome Studies.Appl Environ Microbiol. 2018 Mar 19;84(7):e02627-17. doi: 10.1128/AEM.02627-17. Print 2018 Apr 1. Appl Environ Microbiol. 2018. PMID: 29427429 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

