Virtual screening and experimental validation of novel histone deacetylase inhibitors
- PMID: 27443303
- PMCID: PMC4955146
- DOI: 10.1186/s40360-016-0075-8
Virtual screening and experimental validation of novel histone deacetylase inhibitors
Abstract
Background: Histone deacetylases (HDACs) are promising therapeutic targets for the treatment of cancer, diabetes and other human diseases. HDAC inhibitors, as a new class of potential therapeutic agents, have attracted a great deal of interest for both research and clinical applications. Increasing efforts have been focused on the discovery of HDAC inhibitors and some HDAC inhibitors have been approved for use in cancer therapy. However, most HDAC inhibitors, including the clinically approved agents, do not selectively inhibit the deacetylase activity of class I and II HDAC isforms, and many suffer from metabolic instability. This study aims to identify new HDAC inhibitors by using a high-throughput virtual screening approach.
Methods: An integration of in silico virtual screening and in vitro experimental validation was used to identify novel HDAC inhibitors from a chemical database.
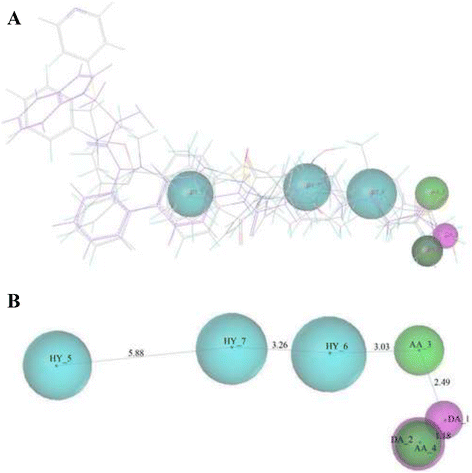
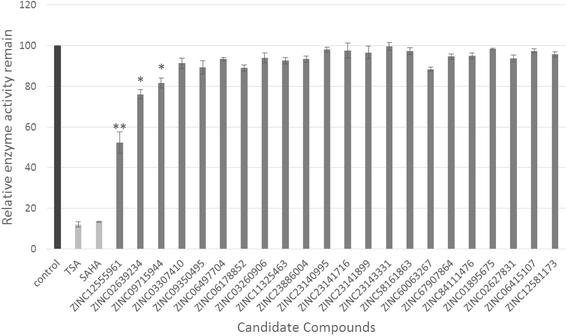
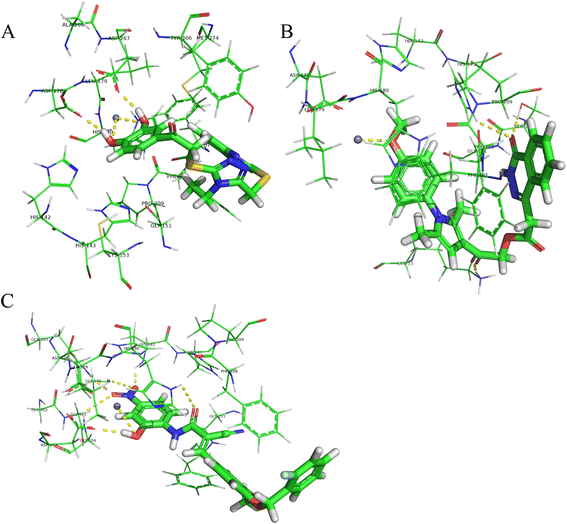
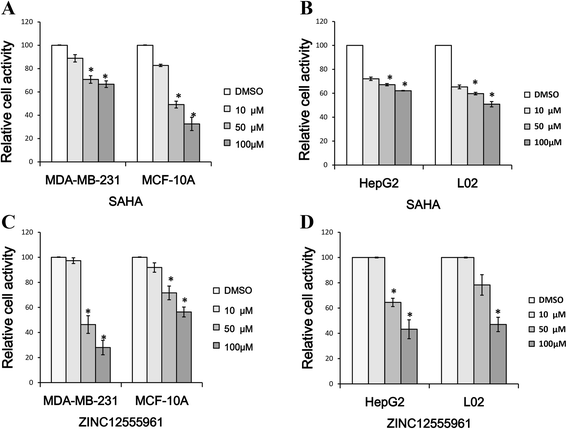
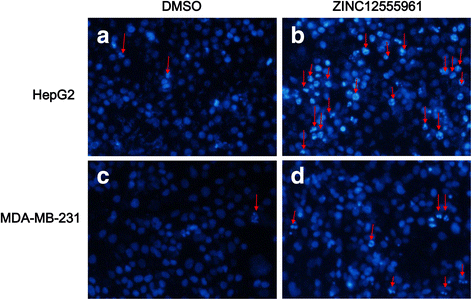
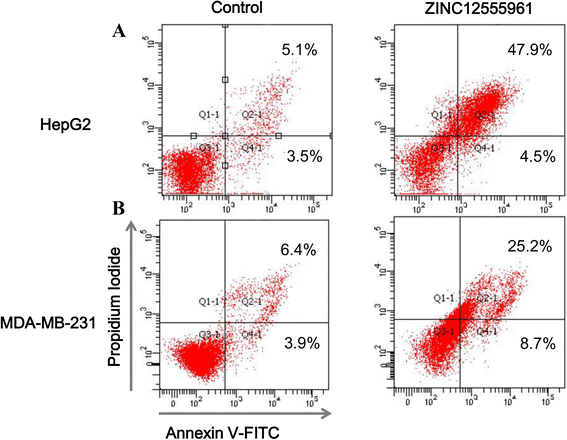
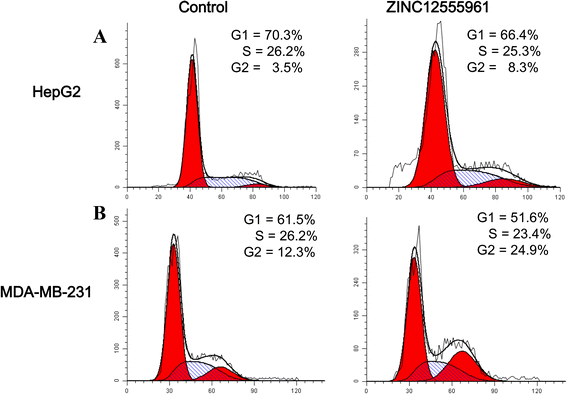
Results: A virtual screening workflow for HDAC inhibitors were created by integrating ligand- and receptor- based virtual screening methods. Using the virtual screening workflow, 22 hit compounds were selected and further tested via in vitro assays. Enzyme inhibition assays showed that three of the 22 compounds had HDAC inhibitory properties. Among these three compounds, ZINC12555961 significantly inhibited HDAC activity. Further in vitro experiments indicated that ZINC12555961 can selectively inhibit proliferation and promote apoptosis of cancer cells.
Conclusions: In summary, our study presents three new and potent HDAC inhibitors and one of these HDAC inhibitors shows anti-proliferative and apoptosis-inducing activity against various cancer cell lines. These results suggest that the developed virtual screening workflow can provide a useful source of information for the screening and validation of new HDAC inhibitors. The new-found HDAC inhibitors are worthy to further and more comprehensive investigations.
Keywords: Apoptosis; Docking; HDAC inhibitors; Pharmacophore; Virtual screening.
Figures
Similar articles
-
An Improved Protocol for the Virtual Screening Discovery of Novel Histone Deacetylase Inhibitors.Chem Pharm Bull (Tokyo). 2019 Oct 1;67(10):1076-1081. doi: 10.1248/cpb.c19-00321. Epub 2019 Aug 9. Chem Pharm Bull (Tokyo). 2019. PMID: 31406093
-
Hierarchical virtual screening of the dual MMP-2/HDAC-6 inhibitors from natural products based on pharmacophore models and molecular docking.J Biomol Struct Dyn. 2019 Feb;37(3):649-670. doi: 10.1080/07391102.2018.1434833. Epub 2018 Feb 12. J Biomol Struct Dyn. 2019. PMID: 29380672
-
Design, synthesis and biological evaluation of novel hydroxamates and 2-aminobenzamides as potent histone deacetylase inhibitors and antitumor agents.Eur J Med Chem. 2017 Jul 7;134:1-12. doi: 10.1016/j.ejmech.2017.03.038. Epub 2017 Mar 22. Eur J Med Chem. 2017. PMID: 28391133
-
In Silico Approach to Finding New Active Compounds from Histone Deacetylase (HDAC) Family.Curr Pharm Des. 2016;22(23):3488-97. doi: 10.2174/1381612822666160414142514. Curr Pharm Des. 2016. PMID: 27075581 Review.
-
Identification of type-specific anticancer histone deacetylase inhibitors: road to success.Cancer Chemother Pharmacol. 2010 Sep;66(4):625-33. doi: 10.1007/s00280-010-1324-y. Epub 2010 Apr 17. Cancer Chemother Pharmacol. 2010. PMID: 20401613 Review.
Cited by
-
Tectorigenin targets PKACα to promote GLUT4 expression in skeletal muscle and improve insulin resistance in vitro and in vivo.Int J Biol Sci. 2023 Mar 5;19(5):1579-1596. doi: 10.7150/ijbs.80125. eCollection 2023. Int J Biol Sci. 2023. PMID: 37056927 Free PMC article.
-
Genome-wide analysis of circular RNAs in bovine cumulus cells treated with BMP15 and GDF9.Sci Rep. 2018 May 21;8(1):7944. doi: 10.1038/s41598-018-26157-2. Sci Rep. 2018. PMID: 29786687 Free PMC article.
-
Exploration of the binding pocket of histone deacetylases: the design of potent and isoform-selective inhibitors.Turk J Biol. 2017 Dec 18;41(6):901-918. doi: 10.3906/biy-1701-26. eCollection 2017. Turk J Biol. 2017. PMID: 30814855 Free PMC article.
-
Discovery of novel N-substituted thiazolidinediones (TZDs) as HDAC8 inhibitors: in-silico studies, synthesis, and biological evaluation.Bioorg Chem. 2020 Jul;100:103934. doi: 10.1016/j.bioorg.2020.103934. Epub 2020 May 15. Bioorg Chem. 2020. PMID: 32446120 Free PMC article.
-
STS1 and STS2 Phosphatase Inhibitor Baicalein Enhances the Expansion of Hematopoietic and Progenitor Stem Cells and Alleviates 5-Fluorouracil-Induced Myelosuppression.Int J Mol Sci. 2023 Feb 3;24(3):2987. doi: 10.3390/ijms24032987. Int J Mol Sci. 2023. PMID: 36769312 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

