Reversion of Cold-Adapted Live Attenuated Influenza Vaccine into a Pathogenic Virus
- PMID: 27440882
- PMCID: PMC5021423
- DOI: 10.1128/JVI.00163-16
Reversion of Cold-Adapted Live Attenuated Influenza Vaccine into a Pathogenic Virus
Abstract
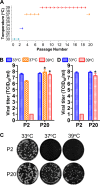
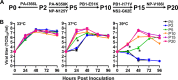
The only licensed live attenuated influenza A virus vaccines (LAIVs) in the United States (FluMist) are created using internal protein-coding gene segments from the cold-adapted temperature-sensitive master donor virus A/Ann Arbor/6/1960 and HA/NA gene segments from circulating viruses. During serial passage of A/Ann Arbor/6/1960 at low temperatures to select the desired attenuating phenotypes, multiple cold-adaptive mutations and temperature-sensitive mutations arose. A substantial amount of scientific and clinical evidence has proven that FluMist is safe and effective. Nevertheless, no study has been conducted specifically to determine if the attenuating temperature-sensitive phenotype can revert and, if so, the types of substitutions that will emerge (i.e., compensatory substitutions versus reversion of existing attenuating mutations). Serial passage of the monovalent FluMist 2009 H1N1 pandemic vaccine at increasing temperatures in vitro generated a variant that replicated efficiently at higher temperatures. Sequencing of the variant identified seven nonsynonymous mutations, PB1-E51K, PB1-I171V, PA-N350K, PA-L366I, NP-N125Y, NP-V186I, and NS2-G63E. None occurred at positions previously reported to affect the temperature sensitivity of influenza A viruses. Synthetic genomics technology was used to synthesize the whole genome of the virus, and the roles of individual mutations were characterized by assessing their effects on RNA polymerase activity and virus replication kinetics at various temperatures. The revertant also regained virulence and caused significant disease in mice, with severity comparable to that caused by a wild-type 2009 H1N1 pandemic virus.
Importance: The live attenuated influenza vaccine FluMist has been proven safe and effective and is widely used in the United States. The phenotype and genotype of the vaccine virus are believed to be very stable, and mutants that cause disease in animals or humans have never been reported. By propagating the virus under well-controlled laboratory conditions, we found that the FluMist vaccine backbone could regain virulence to cause severe disease in mice. The identification of the responsible substitutions and elucidation of the underlying mechanisms provide unique insights into the attenuation of influenza virus, which is important to basic research on vaccines, attenuation reversion, and replication. In addition, this study suggests that the safety of LAIVs should be closely monitored after mass vaccination and that novel strategies to continue to improve LAIV vaccine safety should be investigated.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
Mutation L319Q in the PB1 Polymerase Subunit Improves Attenuation of a Candidate Live-Attenuated Influenza A Virus Vaccine.Microbiol Spectr. 2022 Jun 29;10(3):e0007822. doi: 10.1128/spectrum.00078-22. Epub 2022 May 18. Microbiol Spectr. 2022. PMID: 35583364 Free PMC article.
-
Engineering temperature sensitive live attenuated influenza vaccines from emerging viruses.Vaccine. 2012 May 21;30(24):3691-702. doi: 10.1016/j.vaccine.2012.03.025. Epub 2012 Mar 24. Vaccine. 2012. PMID: 22449422 Free PMC article.
-
The temperature-sensitive and attenuation phenotypes conferred by mutations in the influenza virus PB2, PB1, and NP genes are influenced by the species of origin of the PB2 gene in reassortant viruses derived from influenza A/California/07/2009 and A/WSN/33 viruses.J Virol. 2014 Nov;88(21):12339-47. doi: 10.1128/JVI.02142-14. Epub 2014 Aug 13. J Virol. 2014. PMID: 25122786 Free PMC article.
-
Influenza virus vaccine live intranasal--MedImmune vaccines: CAIV-T, influenza vaccine live intranasal.Drugs R D. 2003;4(5):312-9. doi: 10.2165/00126839-200304050-00007. Drugs R D. 2003. PMID: 12952502 Review.
-
Temperature Sensitive Mutations in Influenza A Viral Ribonucleoprotein Complex Responsible for the Attenuation of the Live Attenuated Influenza Vaccine.Viruses. 2018 Oct 15;10(10):560. doi: 10.3390/v10100560. Viruses. 2018. PMID: 30326610 Free PMC article. Review.
Cited by
-
Development of a Genetically Stable Live Attenuated Influenza Vaccine Strain Using an Engineered High-Fidelity Viral Polymerase.J Virol. 2021 May 24;95(12):e00493-21. doi: 10.1128/JVI.00493-21. Print 2021 May 24. J Virol. 2021. PMID: 33827947 Free PMC article.
-
Dynamic Propagation and Impact of Pandemic Influenza A (2009 H1N1) in Children: A Detailed Review.Curr Microbiol. 2020 Dec;77(12):3809-3820. doi: 10.1007/s00284-020-02213-x. Epub 2020 Sep 21. Curr Microbiol. 2020. PMID: 32959089 Free PMC article. Review.
-
Immune Responses Elicited by Live Attenuated Influenza Vaccines as Correlates of Universal Protection against Influenza Viruses.Vaccines (Basel). 2021 Apr 7;9(4):353. doi: 10.3390/vaccines9040353. Vaccines (Basel). 2021. PMID: 33916924 Free PMC article. Review.
-
Immunization with a suicidal DNA vaccine expressing the E glycoprotein protects ducklings against duck Tembusu virus.Virol J. 2018 Sep 14;15(1):140. doi: 10.1186/s12985-018-1053-0. Virol J. 2018. PMID: 30217161 Free PMC article.
-
Bioengineering Strategies for Developing Vaccines against Respiratory Viral Diseases.Clin Microbiol Rev. 2022 Jan 19;35(1):e0012321. doi: 10.1128/CMR.00123-21. Epub 2021 Nov 17. Clin Microbiol Rev. 2022. PMID: 34788128 Free PMC article. Review.
References
-
- WHO. 2009. World Health Organization: Influenza (seasonal) fact sheet 211. http://www.who.int/mediacentre/factsheets/fs211/en/.
-
- Clements ML, Betts RF, Murphy BR. 1984. Advantage of live attenuated cold-adapted influenza A virus over inactivated vaccine for A/Washington/80 (H3N2) wild-type virus infection. Lancet i:705–708. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

