RpoN Modulates Carbapenem Tolerance in Pseudomonas aeruginosa through Pseudomonas Quinolone Signal and PqsE
- PMID: 27431228
- PMCID: PMC5038263
- DOI: 10.1128/AAC.00260-16
RpoN Modulates Carbapenem Tolerance in Pseudomonas aeruginosa through Pseudomonas Quinolone Signal and PqsE
Abstract
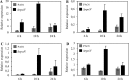
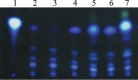
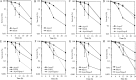
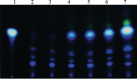
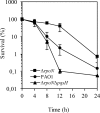
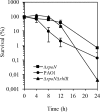
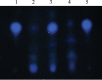
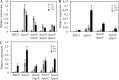
The ability of Pseudomonas aeruginosa to rapidly modulate its response to antibiotic stress and persist in the presence of antibiotics is closely associated with the process of cell-to-cell signaling. The alternative sigma factor RpoN (σ(54)) is involved in the regulation of quorum sensing (QS) and plays an important role in the survival of stationary-phase cells in the presence of carbapenems. Here, we demonstrate that a ΔrpoN mutant grown in nutrient-rich medium has increased expression of pqsA, pqsH, and pqsR throughout growth, resulting in the increased production of the Pseudomonas quinolone signal (PQS). The link between pqsA and its role in carbapenem tolerance was studied using a ΔrpoN ΔpqsA mutant, in which the carbapenem-tolerant phenotype of the ΔrpoN mutant was abolished. In addition, we demonstrate that another mechanism leading to carbapenem tolerance in the ΔrpoN mutant is mediated through pqsE Exogenously supplied PQS abolished the biapenem-sensitive phenotype of the ΔrpoN ΔpqsA mutant, and overexpression of pqsE failed to alter the susceptibility of the ΔrpoN ΔpqsA mutant to biapenem. The mutations in the ΔrpoN ΔrhlR mutant and the ΔrpoN ΔpqsH mutant led to susceptibility to biapenem. Comparison of the changes in the expression of the genes involved in QS in wild-type PAO1 with their expression in the ΔrpoN mutant and the ΔrpoN mutant-derived strains demonstrated the regulatory effect of RpoN on the transcript levels of rhlR, vqsR, and rpoS The findings of this study demonstrate that RpoN negatively regulates the expression of PQS in nutrient-rich medium and provide evidence that RpoN interacts with pqsA, pqsE, pqsH, and rhlR in response to antibiotic stress.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures

Similar articles
-
Role of the interplay between quorum sensing regulator VqsR and the Pseudomonas quinolone signal in mediating carbapenem tolerance in Pseudomonas aeruginosa.Res Microbiol. 2017 Jun;168(5):450-460. doi: 10.1016/j.resmic.2017.02.007. Epub 2017 Mar 2. Res Microbiol. 2017. PMID: 28263907
-
PqsR-independent quorum-sensing response of Pseudomonas aeruginosa ATCC 9027 outlier-strain reveals new insights on the PqsE effect on RhlR activity.Mol Microbiol. 2021 Oct;116(4):1113-1123. doi: 10.1111/mmi.14797. Epub 2021 Aug 30. Mol Microbiol. 2021. PMID: 34418194
-
RpoN Regulates Virulence Factors of Pseudomonas aeruginosa via Modulating the PqsR Quorum Sensing Regulator.Int J Mol Sci. 2015 Nov 30;16(12):28311-9. doi: 10.3390/ijms161226103. Int J Mol Sci. 2015. PMID: 26633362 Free PMC article.
-
The third quorum-sensing system of Pseudomonas aeruginosa: Pseudomonas quinolone signal and the enigmatic PqsE protein.J Med Microbiol. 2020 Jan;69(1):25-34. doi: 10.1099/jmm.0.001116. J Med Microbiol. 2020. PMID: 31794380 Review.
-
Quorum sensing by 2-alkyl-4-quinolones in Pseudomonas aeruginosa and other bacterial species.Mol Biosyst. 2008 Sep;4(9):882-8. doi: 10.1039/b803796p. Epub 2008 Jun 30. Mol Biosyst. 2008. PMID: 18704225 Review.
Cited by
-
Dissemination of Genetic Acquisition/Loss Provides a Variety of Quorum Sensing Regulatory Properties in Pseudoalteromonas.Int J Mol Sci. 2018 Nov 18;19(11):3636. doi: 10.3390/ijms19113636. Int J Mol Sci. 2018. PMID: 30453700 Free PMC article.
-
RpoN Promotes Pseudomonas aeruginosa Survival in the Presence of Tobramycin.Front Microbiol. 2017 May 12;8:839. doi: 10.3389/fmicb.2017.00839. eCollection 2017. Front Microbiol. 2017. PMID: 28553272 Free PMC article.
-
The Regulatory Functions of σ54 Factor in Phytopathogenic Bacteria.Int J Mol Sci. 2021 Nov 24;22(23):12692. doi: 10.3390/ijms222312692. Int J Mol Sci. 2021. PMID: 34884502 Free PMC article. Review.
-
Blocking RpoN reduces virulence of Pseudomonas aeruginosa isolated from cystic fibrosis patients and increases antibiotic sensitivity in a laboratory strain.Sci Rep. 2019 Apr 30;9(1):6677. doi: 10.1038/s41598-019-43060-6. Sci Rep. 2019. PMID: 31040330 Free PMC article.
-
Spatially dependent alkyl quinolone signaling responses to antibiotics in Pseudomonas aeruginosa swarms.J Biol Chem. 2018 Jun 15;293(24):9544-9552. doi: 10.1074/jbc.RA118.002605. Epub 2018 Mar 27. J Biol Chem. 2018. PMID: 29588364 Free PMC article.
References
-
- Hashizume T, Ishino F, Nakagawa J, Tamaki S, Matsuhashi M. 1984. Studies on the mechanism of action of imipenem (N-formimidoylthienamycin) in vitro: binding to the penicillin-binding proteins (PBPs) in Escherichia coli and Pseudomonas aeruginosa, and inhibition of enzyme activities due to the PBPs in E. coli. J Antibiot (Tokyo) 37:394–400. doi:10.7164/antibiotics.37.394. - DOI - PubMed
-
- Monahan LG, Turnbull L, Osvath SR, Birch D, Charles IG, Whitchurch CB. 2014. Rapid conversion of Pseudomonas aeruginosa to a spherical cell morphotype facilitates tolerance to carbapenems and penicillins but increases susceptibility to antimicrobial peptides. Antimicrob Agents Chemother 58:1956–1962. doi:10.1128/AAC.01901-13. - DOI - PMC - PubMed
-
- Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986. doi:10.1126/science.1211037. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

