Human cytomegalovirus miR-UL112-1 promotes the down-regulation of viral immediate early-gene expression during latency to prevent T-cell recognition of latently infected cells
- PMID: 27411311
- PMCID: PMC5756489
- DOI: 10.1099/jgv.0.000546
Human cytomegalovirus miR-UL112-1 promotes the down-regulation of viral immediate early-gene expression during latency to prevent T-cell recognition of latently infected cells
Abstract
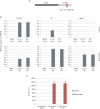
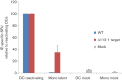
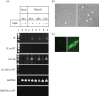
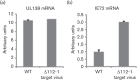
Human cytomegalovirus, a member of the herpesvirus family, can cause significant morbidity and mortality in immune compromised patients resulting from either primary lytic infection or reactivation from latency. Latent infection is associated with a restricted viral transcription programme compared to lytic infection which consists of defined protein coding RNAs but also includes a number of virally encoded microRNAs (miRNAs). One of these, miR-UL112-1, is known to target the major lytic IE72 transcript but, to date, a functional role for miR-UL112-1 during latent infection has not been shown. To address this, we have analysed latent infection in myeloid cells using a virus in which the target site for miR-UL112-1 in the 3' UTR of IE72 was removed such that any IE72 RNA present during latent infection would no longer be subject to regulation by miR-UL112-1 through the RNAi pathway. Our data show that removal of the miR-UL112-1 target site in IE72 results in increased levels of IE72 RNA in experimentally latent primary monocytes. Furthermore, this resulted in induction of immediate early (IE) gene expression that is detectable by IE-specific cytotoxic T-cells (CTLs); no such CTL recognition of monocytes latently infected with wild-type virus was observed. We also recapitulated these findings in the more tractable THP-1 cell line model of latency. These observations argue that an important role for miR-UL112-1 during latency is to ensure tight control of lytic viral immediate early (IE) gene expression thereby preventing recognition of latently infected cells by the host's potent pre-existing anti-viral CTL response.
Figures
Similar articles
-
Human cytomegalovirus latent infection alters the expression of cellular and viral microRNA.Gene. 2014 Feb 25;536(2):272-8. doi: 10.1016/j.gene.2013.12.012. Epub 2013 Dec 18. Gene. 2014. PMID: 24361963
-
Human Cytomegalovirus MicroRNAs miR-US5-1 and miR-UL112-3p Block Proinflammatory Cytokine Production in Response to NF-κB-Activating Factors through Direct Downregulation of IKKα and IKKβ.mBio. 2017 Mar 7;8(2):e00109-17. doi: 10.1128/mBio.00109-17. mBio. 2017. PMID: 28270578 Free PMC article.
-
Latency-Associated Expression of Human Cytomegalovirus US28 Attenuates Cell Signaling Pathways To Maintain Latent Infection.mBio. 2017 Dec 5;8(6):e01754-17. doi: 10.1128/mBio.01754-17. mBio. 2017. PMID: 29208743 Free PMC article.
-
Sleepless latency of human cytomegalovirus.Med Microbiol Immunol. 2015 Jun;204(3):421-9. doi: 10.1007/s00430-015-0401-6. Epub 2015 Mar 14. Med Microbiol Immunol. 2015. PMID: 25772624 Free PMC article. Review.
-
Chromatin-mediated regulation of cytomegalovirus gene expression.Virus Res. 2011 May;157(2):134-43. doi: 10.1016/j.virusres.2010.09.019. Epub 2010 Sep 25. Virus Res. 2011. PMID: 20875471 Free PMC article. Review.
Cited by
-
MicroRNA Regulation of Human Herpesvirus Latency.Viruses. 2022 Jun 2;14(6):1215. doi: 10.3390/v14061215. Viruses. 2022. PMID: 35746686 Free PMC article. Review.
-
Monocytes Latently Infected with Human Cytomegalovirus Evade Neutrophil Killing.iScience. 2019 Feb 22;12:13-26. doi: 10.1016/j.isci.2019.01.007. Epub 2019 Jan 8. iScience. 2019. PMID: 30677738 Free PMC article.
-
Human Cytomegalovirus Host Interactions: EGFR and Host Cell Signaling Is a Point of Convergence Between Viral Infection and Functional Changes in Infected Cells.Front Microbiol. 2021 May 7;12:660901. doi: 10.3389/fmicb.2021.660901. eCollection 2021. Front Microbiol. 2021. PMID: 34025614 Free PMC article. Review.
-
Killer cell proteases can target viral immediate-early proteins to control human cytomegalovirus infection in a noncytotoxic manner.PLoS Pathog. 2020 Apr 13;16(4):e1008426. doi: 10.1371/journal.ppat.1008426. eCollection 2020 Apr. PLoS Pathog. 2020. PMID: 32282833 Free PMC article.
-
Deep Illumina sequencing reveals conserved and novel microRNAs in grass carp in response to grass carp reovirus infection.BMC Genomics. 2017 Feb 20;18(1):195. doi: 10.1186/s12864-017-3562-4. BMC Genomics. 2017. PMID: 28219339 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

