RB1: a prototype tumor suppressor and an enigma
- PMID: 27401552
- PMCID: PMC4949322
- DOI: 10.1101/gad.282145.116
RB1: a prototype tumor suppressor and an enigma
Abstract
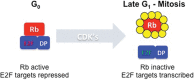
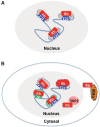
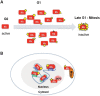
The retinoblastoma susceptibility gene (RB1) was the first tumor suppressor gene to be molecularly defined. RB1 mutations occur in almost all familial and sporadic forms of retinoblastoma, and this gene is mutated at variable frequencies in a variety of other human cancers. Because of its early discovery, the recessive nature of RB1 mutations, and its frequency of inactivation, RB1 is often described as a prototype for the class of tumor suppressor genes. Its gene product (pRB) regulates transcription and is a negative regulator of cell proliferation. Although these general features are well established, a precise description of pRB's mechanism of action has remained elusive. Indeed, in many regards, pRB remains an enigma. This review summarizes some recent developments in pRB research and focuses on progress toward answers for the three fundamental questions that sit at the heart of the pRB literature: What does pRB do? How does the inactivation of RB change the cell? How can our knowledge of RB function be exploited to provide better treatment for cancer patients?
Keywords: E2F; cell proliferation; pRB; tumor suppressor.
© 2016 Dyson; Published by Cold Spring Harbor Laboratory Press.
Figures

Similar articles
-
Phosphorylation of pRb: mechanism for RB pathway inactivation in MYCN-amplified retinoblastoma.Cancer Med. 2017 Mar;6(3):619-630. doi: 10.1002/cam4.1010. Epub 2017 Feb 17. Cancer Med. 2017. PMID: 28211617 Free PMC article.
-
Novel insights into RB1 mutation.Cancer Lett. 2022 Oct 28;547:215870. doi: 10.1016/j.canlet.2022.215870. Epub 2022 Aug 12. Cancer Lett. 2022. PMID: 35964818 Review.
-
Detection of Aberrant DNA Methylation Patterns in the RB1 Gene.Methods Mol Biol. 2018;1726:35-47. doi: 10.1007/978-1-4939-7565-5_5. Methods Mol Biol. 2018. PMID: 29468542
-
The RB1 Story: Characterization and Cloning of the First Tumor Suppressor Gene.Genes (Basel). 2019 Nov 1;10(11):879. doi: 10.3390/genes10110879. Genes (Basel). 2019. PMID: 31683923 Free PMC article. Review.
-
The RASSF6 Tumor Suppressor Protein Regulates Apoptosis and Cell Cycle Progression via Retinoblastoma Protein.Mol Cell Biol. 2018 Aug 15;38(17):e00046-18. doi: 10.1128/MCB.00046-18. Print 2018 Sep 1. Mol Cell Biol. 2018. PMID: 29891515 Free PMC article.
Cited by
-
Biphasic cell cycle defect causes impaired neurogenesis in down syndrome.Front Genet. 2022 Oct 12;13:1007519. doi: 10.3389/fgene.2022.1007519. eCollection 2022. Front Genet. 2022. PMID: 36313423 Free PMC article.
-
Bifidobacterium Strain-Specific Enhances the Efficacy of Cancer Therapeutics in Tumor-Bearing Mice.Cancers (Basel). 2021 Feb 25;13(5):957. doi: 10.3390/cancers13050957. Cancers (Basel). 2021. PMID: 33668827 Free PMC article.
-
Characterizing DNA methylation signatures of retinoblastoma using aqueous humor liquid biopsy.Nat Commun. 2022 Sep 21;13(1):5523. doi: 10.1038/s41467-022-33248-2. Nat Commun. 2022. PMID: 36130950 Free PMC article.
-
Predicting specific mortality from laryngeal cancer based on competing risk model: a retrospective analysis based on the SEER database.Ann Transl Med. 2023 Feb 28;11(4):179. doi: 10.21037/atm-23-400. Ann Transl Med. 2023. PMID: 36923079 Free PMC article.
-
Glioma targeted therapy: insight into future of molecular approaches.Mol Cancer. 2022 Feb 8;21(1):39. doi: 10.1186/s12943-022-01513-z. Mol Cancer. 2022. PMID: 35135556 Free PMC article. Review.
References
-
- Aagaard L, Lukas J, Bartkova J, Kjerulff AA, Strauss M, Bartek J. 1995. Aberrations of p16Ink4 and retinoblastoma tumour-suppressor genes occur in distinct sub-sets of human cancer cell lines. Int J Cancer 61: 115–120. - PubMed
-
- Angus SP, Wheeler LJ, Ranmal SA, Zhang X, Markey MP, Mathews CK, Knudsen ES. 2002. Retinoblastoma tumor suppressor targets dNTP metabolism to regulate DNA replication. J Biol Chem 277: 44376–44384. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
