The transition from linear to highly branched poly(β-amino ester)s: Branching matters for gene delivery
- PMID: 27386572
- PMCID: PMC4928911
- DOI: 10.1126/sciadv.1600102
The transition from linear to highly branched poly(β-amino ester)s: Branching matters for gene delivery
Abstract
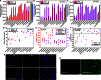
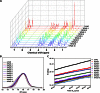
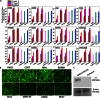
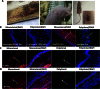
Nonviral gene therapy holds great promise but has not delivered treatments for clinical application to date. Lack of safe and efficient gene delivery vectors is the major hurdle. Among nonviral gene delivery vectors, poly(β-amino ester)s are one of the most versatile candidates because of their wide monomer availability, high polymer flexibility, and superior gene transfection performance both in vitro and in vivo. However, to date, all research has been focused on vectors with a linear structure. A well-accepted view is that dendritic or branched polymers have greater potential as gene delivery vectors because of their three-dimensional structure and multiple terminal groups. Nevertheless, to date, the synthesis of dendritic or branched polymers has been proven to be a well-known challenge. We report the design and synthesis of highly branched poly(β-amino ester)s (HPAEs) via a one-pot "A2 + B3 + C2"-type Michael addition approach and evaluate their potential as gene delivery vectors. We find that the branched structure can significantly enhance the transfection efficiency of poly(β-amino ester)s: Up to an 8521-fold enhancement in transfection efficiency was observed across 12 cell types ranging from cell lines, primary cells, to stem cells, over their corresponding linear poly(β-amino ester)s (LPAEs) and the commercial transfection reagents polyethyleneimine, SuperFect, and Lipofectamine 2000. Moreover, we further demonstrate that HPAEs can correct genetic defects in vivo using a recessive dystrophic epidermolysis bullosa graft mouse model. Our findings prove that the A2 + B3 + C2 approach is highly generalizable and flexible for the design and synthesis of HPAEs, which cannot be achieved by the conventional polymerization approach; HPAEs are more efficient vectors in gene transfection than the corresponding LPAEs. This provides valuable insight into the development and applications of nonviral gene delivery and demonstrates great prospect for their translation to a clinical environment.
Keywords: Gene delivery; RDEB; highly branched polymer; non-viral vector; poly(β-amino ester).
Figures

Similar articles
-
Highly branched poly(β-amino ester)s for skin gene therapy.J Control Release. 2016 Dec 28;244(Pt B):336-346. doi: 10.1016/j.jconrel.2016.06.014. Epub 2016 Jun 8. J Control Release. 2016. PMID: 27288877
-
Highly branched poly(β-amino ester)s for gene delivery in hereditary skin diseases.Adv Drug Deliv Rev. 2021 Sep;176:113842. doi: 10.1016/j.addr.2021.113842. Epub 2021 Jul 20. Adv Drug Deliv Rev. 2021. PMID: 34293384 Review.
-
Efficient and Robust Highly Branched Poly(β-amino ester)/Minicircle COL7A1 Polymeric Nanoparticles for Gene Delivery to Recessive Dystrophic Epidermolysis Bullosa Keratinocytes.ACS Appl Mater Interfaces. 2019 Aug 28;11(34):30661-30672. doi: 10.1021/acsami.9b13135. Epub 2019 Aug 20. ACS Appl Mater Interfaces. 2019. PMID: 31390173
-
Tailoring highly branched poly(β-amino ester)s: a synthetic platform for epidermal gene therapy.Chem Commun (Camb). 2015 May 18;51(40):8473-6. doi: 10.1039/c5cc02193f. Chem Commun (Camb). 2015. PMID: 25892461
-
Michael Addition Polymerization of Trifunctional Amine and Acrylic Monomer: A Versatile Platform for Development of Biomaterials.Biomacromolecules. 2016 Oct 10;17(10):3115-3126. doi: 10.1021/acs.biomac.6b01043. Epub 2016 Sep 15. Biomacromolecules. 2016. PMID: 27599254 Review.
Cited by
-
Non-viral delivery of CRISPR-Cas9 complexes for targeted gene editing via a polymer delivery system.Gene Ther. 2022 Apr;29(3-4):157-170. doi: 10.1038/s41434-021-00282-6. Epub 2021 Aug 6. Gene Ther. 2022. PMID: 34363036 Free PMC article.
-
Complex polymer architectures through free-radical polymerization of multivinyl monomers.Nat Rev Chem. 2020 Apr;4(4):194-212. doi: 10.1038/s41570-020-0170-7. Epub 2020 Mar 23. Nat Rev Chem. 2020. PMID: 37128047 Review.
-
Systematic Investigation of Biocompatible Cationic Polymeric Nucleic Acid Carriers for Immunotherapy of Hepatocellular Carcinoma.Cancers (Basel). 2021 Dec 24;14(1):85. doi: 10.3390/cancers14010085. Cancers (Basel). 2021. PMID: 35008249 Free PMC article. Review.
-
Genome-editing prodrug: Targeted delivery and conditional stabilization of CRISPR-Cas9 for precision therapy of inflammatory disease.Sci Adv. 2021 Dec 10;7(50):eabj0624. doi: 10.1126/sciadv.abj0624. Epub 2021 Dec 8. Sci Adv. 2021. PMID: 34878850 Free PMC article.
-
Delivery of siRNA therapeutics using cowpea chlorotic mottle virus-like particles.Biomater Sci. 2019 Aug 1;7(8):3138-3142. doi: 10.1039/c9bm00785g. Epub 2019 Jul 1. Biomater Sci. 2019. PMID: 31257379 Free PMC article.
References
-
- Pack D. W., Hoffman A. S., Pun S., Stayton P. S., Design and development of polymers for gene delivery. Nat. Rev. Drug Discov. 4, 581–593 (2005). - PubMed
-
- Dahlman J. E., Barnes C., Khan O. F., Thiriot A., Jhunjunwala S., Shaw T. E., Xing Y., Sager H. B., Sahay G., Speciner L., Bader A., Bogorad R. L., Yin H., Racie T., Dong Y., Jiang S., Seedorf D., Dave A., Singh Sandhu K., Webber M. J., Novobrantseva T., Ruda V. M., Lytton-Jean A. K. R., Levins C. G., Kalish B., Mudge D. K., Perez M., Abezgauz L., Dutta P., Smith L., Charisse K., Kieran M. W., Fitzgerald K., Nahrendorf M., Danino D., Tuder R. M., von Adrian U. H., Akinc A., Panigrahy D., Schroeder A., Koteliansky V., Langer R., Anderson D. G., In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. Nat. Nanotechnol. 9, 648–655 (2014). - PMC - PubMed
-
- Minter M. A., Simanek E. E., Nonviral vectors for gene delivery. Chem. Rev. 109, 259–302 (2009). - PubMed
-
- Zhao T., Zhang H., Newland B., Aied A., Zhou D., Wang W., Significance of branching for transfection: Synthesis of highly branched degradable functional poly(dimethylaminoethyl methacrylate) by vinyl oligomer combination. Angew. Chem. Int. Ed. Engl. 53, 6095–6100 (2014). - PubMed
-
- Luo D., Saltzman M., Synthetic DNA delivery systems. Nat. Biotechnol. 18, 33–37 (2000). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

