The role of heparan sulphate in development: the ectodermal story
- PMID: 27385054
- PMCID: PMC4960573
- DOI: 10.1111/iep.12180
The role of heparan sulphate in development: the ectodermal story
Abstract
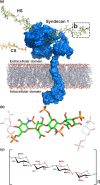
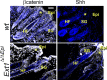
Heparan sulphate (HS) is ubiquitously expressed and is formed of repeating glucosamine and glucuronic/iduronic acid units which are generally highly sulphated. HS is found in tissues bound to proteins forming HS proteoglycans (HSPGs) which are present on the cell membrane or in the extracellular matrix. HSPGs influence a variety of biological processes by interacting with physiologically important proteins, such as morphogens, creating storage pools, generating morphogen gradients and directly mediating signalling pathways, thereby playing vital roles during development. This review discusses the vital role HS plays in the development of tissues from the ectodermal lineage. The ectodermal layer differentiates to form the nervous system (including the spine, peripheral nerves and brain), eye, epidermis, skin appendages and tooth enamel.
© 2016 The Authors. International Journal of Experimental Pathology © 2016 International Journal of Experimental Pathology.
Figures


Similar articles
-
Mesenchymal stem cells, neural lineage potential, heparan sulfate proteoglycans and the matrix.Dev Biol. 2014 Apr 1;388(1):1-10. doi: 10.1016/j.ydbio.2014.01.024. Epub 2014 Feb 6. Dev Biol. 2014. PMID: 24509075 Review.
-
Heparan Sulfate in the Tumor Microenvironment.Adv Exp Med Biol. 2020;1245:147-161. doi: 10.1007/978-3-030-40146-7_7. Adv Exp Med Biol. 2020. PMID: 32266657 Review.
-
Heparan sulfate proteoglycans including syndecan-3 modulate BMP activity during limb cartilage differentiation.Matrix Biol. 2006 Jan;25(1):27-39. doi: 10.1016/j.matbio.2005.07.008. Epub 2005 Oct 13. Matrix Biol. 2006. PMID: 16226436
-
Implications of heparan sulfate and heparanase in neuroinflammation.Matrix Biol. 2014 Apr;35:174-81. doi: 10.1016/j.matbio.2013.12.009. Epub 2014 Jan 4. Matrix Biol. 2014. PMID: 24398134 Review.
-
Heparan sulfate in the nucleus and its control of cellular functions.Matrix Biol. 2014 Apr;35:56-9. doi: 10.1016/j.matbio.2013.10.009. Epub 2013 Dec 3. Matrix Biol. 2014. PMID: 24309018 Free PMC article. Review.
Cited by
-
Syndecan-3 contributes to the regulation of the microenvironment at the node of Ranvier following end-to‑side neurorrhaphy: sodium image analysis.Histochem Cell Biol. 2021 Mar;155(3):355-367. doi: 10.1007/s00418-020-01936-z. Epub 2020 Nov 10. Histochem Cell Biol. 2021. PMID: 33170350
-
Chondroitin Sulphate/Dermatan Sulphate Proteoglycans: Potential Regulators of Corneal Stem/Progenitor Cell Phenotype In Vitro.Int J Mol Sci. 2023 Jan 20;24(3):2095. doi: 10.3390/ijms24032095. Int J Mol Sci. 2023. PMID: 36768414 Free PMC article.
-
Importance of Heparan Sulfate Proteoglycans in Pancreatic Islets and β-Cells.Int J Mol Sci. 2022 Oct 11;23(20):12082. doi: 10.3390/ijms232012082. Int J Mol Sci. 2022. PMID: 36292936 Free PMC article. Review.
-
Epigenetic Regulation of the Biosynthesis & Enzymatic Modification of Heparan Sulfate Proteoglycans: Implications for Tumorigenesis and Cancer Biomarkers.Int J Mol Sci. 2017 Jun 26;18(7):1361. doi: 10.3390/ijms18071361. Int J Mol Sci. 2017. PMID: 28672878 Free PMC article. Review.
-
HS, an Ancient Molecular Recognition and Information Storage Glycosaminoglycan, Equips HS-Proteoglycans with Diverse Matrix and Cell-Interactive Properties Operative in Tissue Development and Tissue Function in Health and Disease.Int J Mol Sci. 2023 Jan 6;24(2):1148. doi: 10.3390/ijms24021148. Int J Mol Sci. 2023. PMID: 36674659 Free PMC article. Review.
References
-
- Ai X., Kitazawa T., Do A.T., Kusche‐Gullberg M., Labosky P.A. & Emerson C.P. Jr (2007) SULF1 and SULF2 regulate heparan sulfate‐mediated GDNF signaling for esophageal innervation. Development 134, 3327–3338. - PubMed
-
- Aikawa J. & Esko J.D. (1999) Molecular cloning and expression of a third member of the heparan sulfate/heparin GlcNAc N‐deacetylase/N‐sulfotransferase family. J. Biol. Chem. 274, 2690–2695. - PubMed
-
- Arikawa‐Hirasawa E., Watanabe H., Takami H., Hassell J.R. & Yamada Y. (1999) Perlecan is essential for cartilage and cephalic development. Nat. Genet. 23, 354–358. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

