Selective Deletion of Astroglial FMRP Dysregulates Glutamate Transporter GLT1 and Contributes to Fragile X Syndrome Phenotypes In Vivo
- PMID: 27383586
- PMCID: PMC4938857
- DOI: 10.1523/JNEUROSCI.1069-16.2016
Selective Deletion of Astroglial FMRP Dysregulates Glutamate Transporter GLT1 and Contributes to Fragile X Syndrome Phenotypes In Vivo
Abstract
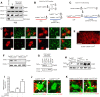
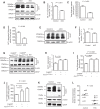
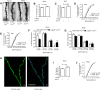
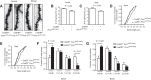
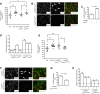
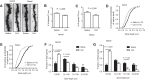
How the loss of fragile X mental retardation protein (FMRP) in different brain cell types, especially in non-neuron glial cells, induces fragile X syndrome (FXS) phenotypes has just begun to be understood. In the current study, we generated inducible astrocyte-specific Fmr1 conditional knock-out mice (i-astro-Fmr1-cKO) and restoration mice (i-astro-Fmr1-cON) to study the in vivo modulation of FXS synaptic phenotypes by astroglial FMRP. We found that functional expression of glutamate transporter GLT1 is 40% decreased in i-astro-Fmr1-cKO somatosensory cortical astrocytes in vivo, which can be fully rescued by the selective re-expression of FMRP in astrocytes in i-astro-Fmr1-cON mice. Although the selective loss of astroglial FMRP only modestly increases spine density and length in cortical pyramidal neurons, selective re-expression of FMRP in astrocytes significantly attenuates abnormal spine morphology in these neurons of i-astro-Fmr1-cON mice. Moreover, we found that basal protein synthesis levels and immunoreactivity of phosphorylated S6 ribosomal protein (p-s6P) is significantly increased in i-astro-Fmr1-cKO mice, while the enhanced cortical protein synthesis observed in Fmr1 KO mice is mitigated in i-astro-Fmr1-cON mice. Furthermore, ceftriaxone-mediated upregulation of surface GLT1 expression restores functional glutamate uptake and attenuates enhanced neuronal excitability in Fmr1 KO mice. In particular, ceftriaxone significantly decreases the growth rate of abnormally accelerated body weight and completely corrects spine abnormality in Fmr1 KO mice. Together, these results show that the selective loss of astroglial FMRP contributes to cortical synaptic deficits in FXS, presumably through dysregulated astroglial glutamate transporter GLT1 and impaired glutamate uptake. These results suggest the involvement of astrocyte-mediated mechanisms in the pathogenesis of FXS.
Significance statement: Previous studies to understand how the loss of function of fragile X mental retardation protein (FMRP) causes fragile X syndrome (FXS) have largely focused on neurons; whether the selective loss of astroglial FMRP in vivo alters astrocyte functions and contributes to the pathogenesis of FXS remain essentially unknown. This has become a long-standing unanswered question in the fragile X field, which is also relevant to autism pathogenesis. Our current study generated astrocyte-specific Fmr1 conditional knock-out and restoration mice, and provided compelling evidence that the selective loss of astroglial FMRP contributes to cortical synaptic deficits in FXS, likely through the dysregulated astroglial glutamate transporter GLT1 expression and impaired glutamate uptake. These results demonstrate previously undescribed astrocyte-mediated mechanisms in the pathogenesis of FXS.
Keywords: FMRP; astrocyte; autism; fragile X; glutamate transporter; protein synthesis.
Copyright © 2016 the authors 0270-6474/16/367080-16$15.00/0.
Figures


Similar articles
-
Astroglial FMRP-dependent translational down-regulation of mGluR5 underlies glutamate transporter GLT1 dysregulation in the fragile X mouse.Hum Mol Genet. 2013 May 15;22(10):2041-54. doi: 10.1093/hmg/ddt055. Epub 2013 Feb 7. Hum Mol Genet. 2013. PMID: 23396537 Free PMC article.
-
Astroglial FMRP modulates synaptic signaling and behavior phenotypes in FXS mouse model.Glia. 2021 Mar;69(3):594-608. doi: 10.1002/glia.23915. Epub 2020 Sep 24. Glia. 2021. PMID: 32970902
-
Deletion of Fmr1 in parvalbumin-expressing neurons results in dysregulated translation and selective behavioral deficits associated with fragile X syndrome.Mol Autism. 2022 Jun 29;13(1):29. doi: 10.1186/s13229-022-00509-2. Mol Autism. 2022. PMID: 35768828 Free PMC article.
-
BDNF in fragile X syndrome.Neuropharmacology. 2014 Jan;76 Pt C:729-36. doi: 10.1016/j.neuropharm.2013.05.018. Epub 2013 May 29. Neuropharmacology. 2014. PMID: 23727436 Review.
-
Fragile X syndrome: From protein function to therapy.Am J Med Genet A. 2013 Nov;161A(11):2809-21. doi: 10.1002/ajmg.a.36241. Epub 2013 Sep 24. Am J Med Genet A. 2013. PMID: 24115651 Review.
Cited by
-
Channelopathies in fragile X syndrome.Nat Rev Neurosci. 2021 May;22(5):275-289. doi: 10.1038/s41583-021-00445-9. Epub 2021 Apr 7. Nat Rev Neurosci. 2021. PMID: 33828309 Free PMC article. Review.
-
Neuron-Astrocyte Interactions: A Human Perspective.Adv Neurobiol. 2024;39:69-93. doi: 10.1007/978-3-031-64839-7_4. Adv Neurobiol. 2024. PMID: 39190072 Review.
-
Dysregulated Purinergic Signalling in Fragile X Syndrome Cortical Astrocytes.Neuromolecular Med. 2024 Sep 10;26(1):36. doi: 10.1007/s12017-024-08802-4. Neuromolecular Med. 2024. PMID: 39254908
-
Conditional Knock-out of mGluR5 from Astrocytes during Epilepsy Development Impairs High-Frequency Glutamate Uptake.J Neurosci. 2019 Jan 23;39(4):727-742. doi: 10.1523/JNEUROSCI.1148-18.2018. Epub 2018 Nov 30. J Neurosci. 2019. PMID: 30504280 Free PMC article.
-
The Regulation of Astrocytic Glutamate Transporters in Health and Neurodegenerative Diseases.Int J Mol Sci. 2020 Dec 17;21(24):9607. doi: 10.3390/ijms21249607. Int J Mol Sci. 2020. PMID: 33348528 Free PMC article. Review.
References
-
- Bakker CE, Verheij C, Willemsen R, van der Helm R, Oerlemans F, Vermey M, Bygrave A, Hoogeveen AT, Oostra BA, Reyniers E, D'Hooge R, Cras P, Van Velzen D, Nagels G, Martin JJ, De Deyn PP, Darby JK, Willems PJ. Fmr1 knockout mice: a model to study fragile X mental retardation. Cell. 1994;78:23–33. - PubMed
-
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
