Long non-coding RNA expression profiling in the NCI60 cancer cell line panel using high-throughput RT-qPCR
- PMID: 27377824
- PMCID: PMC4932877
- DOI: 10.1038/sdata.2016.52
Long non-coding RNA expression profiling in the NCI60 cancer cell line panel using high-throughput RT-qPCR
Abstract
Long non-coding RNAs (lncRNAs) form a new class of RNA molecules implicated in various aspects of protein coding gene expression regulation. To study lncRNAs in cancer, we generated expression profiles for 1707 human lncRNAs in the NCI60 cancer cell line panel using a high-throughput nanowell RT-qPCR platform. We describe how qPCR assays were designed and validated and provide processed and normalized expression data for further analysis. Data quality is demonstrated by matching the lncRNA expression profiles with phenotypic and genomic characteristics of the cancer cell lines. This data set can be integrated with publicly available omics and pharmacological data sets to uncover novel associations between lncRNA expression and mRNA expression, miRNA expression, DNA copy number, protein coding gene mutation status or drug response.
Conflict of interest statement
Primer sequences are property of Biogazelle and non-exclusively licensed to WaferGen Biosystems.
Figures
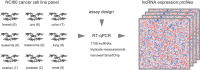
Comment on
-
Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study.Nat Methods. 2014 Aug;11(8):809-15. doi: 10.1038/nmeth.3014. Epub 2014 Jun 29. Nat Methods. 2014. PMID: 24973947
-
Melanoma addiction to the long non-coding RNA SAMMSON.Nature. 2016 Mar 24;531(7595):518-22. doi: 10.1038/nature17161. Nature. 2016. PMID: 27008969
Similar articles
-
Transcriptome profiling of lncRNA and co-expression networks in esophageal squamous cell carcinoma by RNA sequencing.Tumour Biol. 2016 Oct;37(10):13091-13100. doi: 10.1007/s13277-016-5227-3. Epub 2016 Jul 23. Tumour Biol. 2016. PMID: 27449043
-
Identification of crucial regulatory relationships between long non-coding RNAs and protein-coding genes in lung squamous cell carcinoma.Mol Cell Probes. 2016 Jun;30(3):146-52. doi: 10.1016/j.mcp.2016.02.009. Epub 2016 Feb 27. Mol Cell Probes. 2016. PMID: 26928440
-
Aberrantly expressed long noncoding RNAs in recurrent implantation failure: A microarray related study.Syst Biol Reprod Med. 2017 Aug;63(4):269-278. doi: 10.1080/19396368.2017.1310329. Epub 2017 Apr 25. Syst Biol Reprod Med. 2017. PMID: 28441042
-
Identification of Gossypium hirsutum long non-coding RNAs (lncRNAs) under salt stress.BMC Plant Biol. 2018 Jan 25;18(1):23. doi: 10.1186/s12870-018-1238-0. BMC Plant Biol. 2018. PMID: 29370759 Free PMC article.
-
Perspectives of long non-coding RNAs in cancer.Mol Biol Rep. 2017 Apr;44(2):203-218. doi: 10.1007/s11033-017-4103-6. Epub 2017 Apr 8. Mol Biol Rep. 2017. PMID: 28391434 Review.
Cited by
-
lncRNASNP v3: an updated database for functional variants in long non-coding RNAs.Nucleic Acids Res. 2023 Jan 6;51(D1):D192-D198. doi: 10.1093/nar/gkac981. Nucleic Acids Res. 2023. PMID: 36350671 Free PMC article.
References
Data Citations
-
- Mestdagh P., Vandesompele J. 2016. Gene Expression Omnibus. GSE80332
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

