Structural effects of modified ribonucleotides and magnesium in transfer RNAs
- PMID: 27364608
- PMCID: PMC5052103
- DOI: 10.1016/j.bmc.2016.06.037
Structural effects of modified ribonucleotides and magnesium in transfer RNAs
Abstract
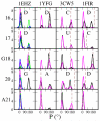
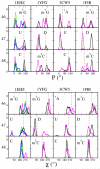
Modified nucleotides are ubiquitous and important to tRNA structure and function. To understand their effect on tRNA conformation, we performed a series of molecular dynamics simulations on yeast tRNAPhe and tRNAinit, Escherichia coli tRNAinit and HIV tRNALys. Simulations were performed with the wild type modified nucleotides, using the recently developed CHARMM compatible force field parameter set for modified nucleotides (J. Comput. Chem.2016, 37, 896), or with the corresponding unmodified nucleotides, and in the presence or absence of Mg2+. Results showed a stabilizing effect associated with the presence of the modifications and Mg2+ for some important positions, such as modified guanosine in position 37 and dihydrouridines in 16/17 including both structural properties and base interactions. Some other modifications were also found to make subtle contributions to the structural properties of local domains. While we were not able to investigate the effect of adenosine 37 in tRNAinit and limitations were observed in the conformation of E. coli tRNAinit, the presence of the modified nucleotides and of Mg2+ better maintained the structural features and base interactions of the tRNA systems than in their absence indicating the utility of incorporating the modified nucleotides in simulations of tRNA and other RNAs.
Keywords: Magnesium; Modified nucleotide; Molecular dynamics; Nucleic acids; Structure; Transfer RNA.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures
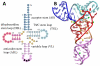
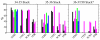

Similar articles
-
Mechanism, specificity and general properties of the yeast enzyme catalysing the formation of inosine 34 in the anticodon of transfer RNA.J Mol Biol. 1996 Oct 4;262(4):437-58. doi: 10.1006/jmbi.1996.0527. J Mol Biol. 1996. PMID: 8893855
-
Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: the effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A.Biochemistry. 2005 Jun 7;44(22):8078-89. doi: 10.1021/bi050343f. Biochemistry. 2005. PMID: 15924427
-
Solution conformations of unmodified and A(37)N(6)-dimethylallyl modified anticodon stem-loops of Escherichia coli tRNA(Phe).J Mol Biol. 2002 Jun 21;319(5):1015-34. doi: 10.1016/S0022-2836(02)00382-0. J Mol Biol. 2002. PMID: 12079344
-
High-resolution nuclear magnetic resonance investigations of the structure of tRNA in solution.Prog Nucleic Acid Res Mol Biol. 1976;18:91-149. doi: 10.1016/s0079-6603(08)60587-5. Prog Nucleic Acid Res Mol Biol. 1976. PMID: 790475 Review. No abstract available.
-
Ribosomal binding of modified tRNA anticodons related to thermal stability.Nucleic Acids Symp Ser. 1997;(36):58-60. Nucleic Acids Symp Ser. 1997. PMID: 9478206 Review.
Cited by
-
Methylation modifications in tRNA and associated disorders: Current research and potential therapeutic targets.Cell Prolif. 2024 Sep;57(9):e13692. doi: 10.1111/cpr.13692. Epub 2024 Jun 28. Cell Prolif. 2024. PMID: 38943267 Free PMC article. Review.
-
tRNA Modifications and Modifying Enzymes in Disease, the Potential Therapeutic Targets.Int J Biol Sci. 2023 Feb 13;19(4):1146-1162. doi: 10.7150/ijbs.80233. eCollection 2023. Int J Biol Sci. 2023. PMID: 36923941 Free PMC article. Review.
-
The Importance of Being Modified: The Role of RNA Modifications in Translational Fidelity.Enzymes. 2017;41:1-50. doi: 10.1016/bs.enz.2017.03.005. Epub 2017 Apr 22. Enzymes. 2017. PMID: 28601219 Free PMC article. Review.
References
-
- Carell T, Brandmayr C, Hienzsch A, Muller M, Pearson D, Reiter V, Thoma I, Thumbs P, Wagner M. Structure and function of noncanonical nucleobases. Angew Chem Int Ed Engl. 2012;51:7110–7131. - PubMed
-
- Giege R, Juhling F, Putz J, Stadler P, Sauter C, Florentz C. Structure of transfer RNAs: similarity and variability. Wiley Interdisciplinary Reviews-Rna. 2012;3:37–61. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

