Phosphoproteome and Transcriptome of RA-Responsive and RA-Resistant Breast Cancer Cell Lines
- PMID: 27362937
- PMCID: PMC4928811
- DOI: 10.1371/journal.pone.0157290
Phosphoproteome and Transcriptome of RA-Responsive and RA-Resistant Breast Cancer Cell Lines
Abstract
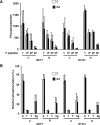
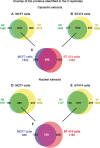
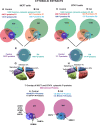
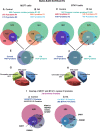
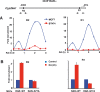
Retinoic acid (RA), the main active vitamin A metabolite, controls multiple biological processes such as cell proliferation and differentiation through genomic programs and kinase cascades activation. Due to these properties, RA has proven anti-cancer capacity. Several breast cancer cells respond to the antiproliferative effects of RA, while others are RA-resistant. However, the overall signaling and transcriptional pathways that are altered in such cells have not been elucidated. Here, in a large-scale analysis of the phosphoproteins and in a genome-wide analysis of the RA-regulated genes, we compared two human breast cancer cell lines, a RA-responsive one, the MCF7 cell line, and a RA-resistant one, the BT474 cell line, which depicts several alterations of the "kinome". Using high-resolution nano-LC-LTQ-Orbitrap mass spectrometry associated to phosphopeptide enrichment, we found that several proteins involved in signaling and in transcription, are differentially phosphorylated before and after RA addition. The paradigm of these proteins is the RA receptor α (RARα), which was phosphorylated in MCF7 cells but not in BT474 cells after RA addition. The panel of the RA-regulated genes was also different. Overall our results indicate that RA resistance might correlate with the deregulation of the phosphoproteome with consequences on gene expression.
Conflict of interest statement
Figures




Similar articles
-
Comparison of N-(4-hydroxyphenyl)retinamide and all-trans-retinoic acid in the regulation of retinoid receptor-mediated gene expression in human breast cancer cell lines.Cancer Res. 1996 Mar 1;56(5):1056-62. Cancer Res. 1996. PMID: 8640761
-
Retinoic acid induces expression of the interleukin-1beta gene in cultured normal human mammary epithelial cells and in human breast carcinoma lines.J Cell Physiol. 2002 Nov;193(2):244-52. doi: 10.1002/jcp.10173. J Cell Physiol. 2002. PMID: 12385002
-
Retinoic acid increases tyrosine phosphorylation of focal adhesion kinase and paxillin in MCF-7 human breast cancer cells.Cancer Res. 1999 Jan 1;59(1):85-90. Cancer Res. 1999. PMID: 9892191
-
The p85α regulatory subunit of PI3K mediates cAMP-PKA and retinoic acid biological effects on MCF7 cell growth and migration.Int J Oncol. 2012 May;40(5):1627-35. doi: 10.3892/ijo.2012.1383. Epub 2012 Feb 21. Int J Oncol. 2012. PMID: 22366926
-
Retinoic acid-induced growth arrest of MCF-7 cells involves the selective regulation of the IRS-1/PI 3-kinase/AKT pathway.Oncogene. 2003 May 29;22(22):3353-60. doi: 10.1038/sj.onc.1206485. Oncogene. 2003. PMID: 12776186
Cited by
-
Design and characterization of mutant and wildtype huntingtin proteins produced from a toolkit of scalable eukaryotic expression systems.J Biol Chem. 2019 Apr 26;294(17):6986-7001. doi: 10.1074/jbc.RA118.007204. Epub 2019 Mar 6. J Biol Chem. 2019. PMID: 30842263 Free PMC article.
-
Cellular retinoid binding-proteins, CRBP, CRABP, FABP5: Effects on retinoid metabolism, function and related diseases.Pharmacol Ther. 2017 May;173:19-33. doi: 10.1016/j.pharmthera.2017.01.004. Epub 2017 Jan 27. Pharmacol Ther. 2017. PMID: 28132904 Free PMC article. Review.
-
Prediction of protein-DNA interactions of transcription factors linking proteomics and transcriptomics data.EuPA Open Proteom. 2016 Sep 15;13:14-23. doi: 10.1016/j.euprot.2016.09.001. eCollection 2016 Dec. EuPA Open Proteom. 2016. PMID: 29900118 Free PMC article.
-
A Fresh Look at the Structure, Regulation, and Functions of Fodrin.Mol Cell Biol. 2020 Aug 14;40(17):e00133-20. doi: 10.1128/MCB.00133-20. Print 2020 Aug 14. Mol Cell Biol. 2020. PMID: 32601107 Free PMC article. Review.
-
The phospho-landscape of the survival of motoneuron protein (SMN) protein: relevance for spinal muscular atrophy (SMA).Cell Mol Life Sci. 2022 Aug 25;79(9):497. doi: 10.1007/s00018-022-04522-9. Cell Mol Life Sci. 2022. PMID: 36006469 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

