Profiling genome-wide DNA methylation
- PMID: 27358654
- PMCID: PMC4926291
- DOI: 10.1186/s13072-016-0075-3
Profiling genome-wide DNA methylation
Abstract
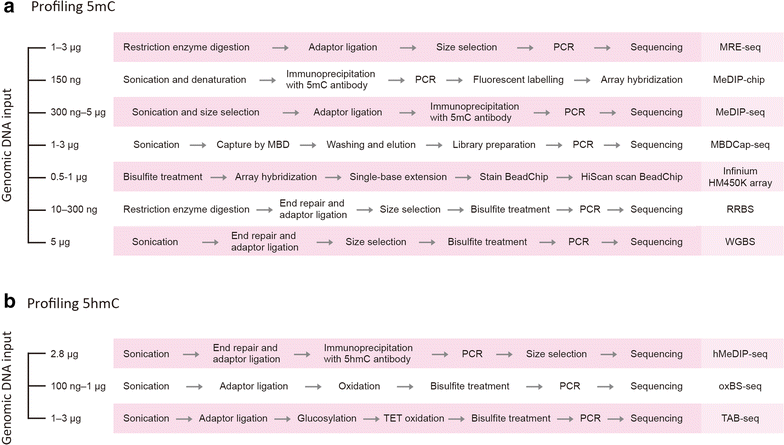
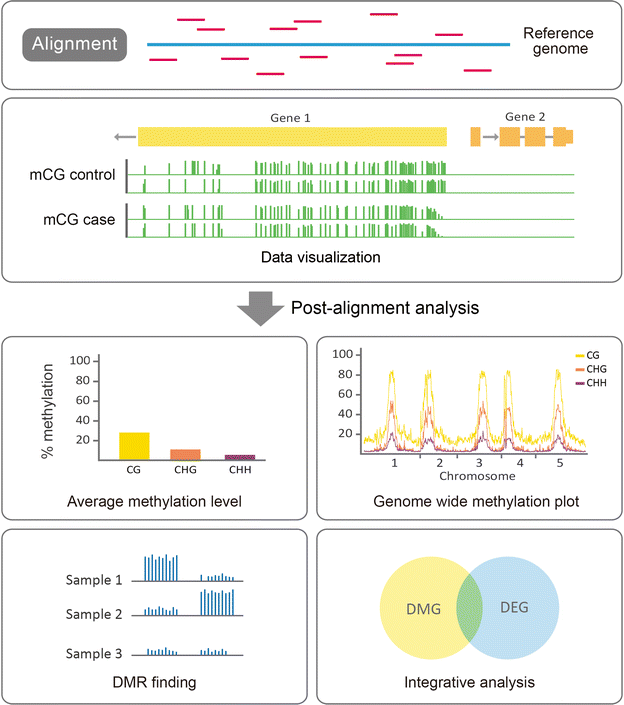
DNA methylation is an epigenetic modification that plays an important role in regulating gene expression and therefore a broad range of biological processes and diseases. DNA methylation is tissue-specific, dynamic, sequence-context-dependent and trans-generationally heritable, and these complex patterns of methylation highlight the significance of profiling DNA methylation to answer biological questions. In this review, we surveyed major methylation assays, along with comparisons and biological examples, to provide an overview of DNA methylation profiling techniques. The advances in microarray and sequencing technologies make genome-wide profiling possible at a single-nucleotide or even a single-cell resolution. These profiling approaches vary in many aspects, such as DNA input, resolution, genomic region coverage, and bioinformatics analysis, and selecting a feasible method requires knowledge of these methods. We first introduce the biological background of DNA methylation and its pattern in plants, animals and fungi. We present an overview of major experimental approaches to profiling genome-wide DNA methylation and hydroxymethylation and then extend to the single-cell methylome. To evaluate these methods, we outline their strengths and weaknesses and perform comparisons across the different platforms. Due to the increasing need to compute high-throughput epigenomic data, we interrogate the computational pipeline for bisulfite sequencing data and also discuss the concept of identifying differentially methylated regions (DMRs). This review summarizes the experimental and computational concepts for profiling genome-wide DNA methylation, followed by biological examples. Overall, this review provides researchers useful guidance for the selection of a profiling method suited to specific research questions.
Keywords: Bisulfite sequencing; DNA methylation; Hydroxymethylation; Methylome; RRBS; Single-cell; WGBS.
Figures

Similar articles
-
Plant-RRBS, a bisulfite and next-generation sequencing-based methylome profiling method enriching for coverage of cytosine positions.BMC Plant Biol. 2017 Jul 6;17(1):115. doi: 10.1186/s12870-017-1070-y. BMC Plant Biol. 2017. PMID: 28683715 Free PMC article.
-
Combining MeDIP-seq and MRE-seq to investigate genome-wide CpG methylation.Methods. 2015 Jan 15;72:29-40. doi: 10.1016/j.ymeth.2014.10.032. Epub 2014 Nov 6. Methods. 2015. PMID: 25448294 Free PMC article.
-
Global DNA methylation profiling technologies and the ovarian cancer methylome.Methods Mol Biol. 2015;1238:653-75. doi: 10.1007/978-1-4939-1804-1_34. Methods Mol Biol. 2015. PMID: 25421685 Review.
-
Tools and Strategies for Analysis of Genome-Wide and Gene-Specific DNA Methylation Patterns.Methods Mol Biol. 2017;1537:249-277. doi: 10.1007/978-1-4939-6685-1_15. Methods Mol Biol. 2017. PMID: 27924599 Review.
-
Profiling DNA Methylation Based on Next-Generation Sequencing Approaches: New Insights and Clinical Applications.Genes (Basel). 2018 Aug 23;9(9):429. doi: 10.3390/genes9090429. Genes (Basel). 2018. PMID: 30142958 Free PMC article. Review.
Cited by
-
CGGBP1-regulated cytosine methylation at CTCF-binding motifs resists stochasticity.BMC Genet. 2020 Jul 29;21(1):84. doi: 10.1186/s12863-020-00894-8. BMC Genet. 2020. PMID: 32727353 Free PMC article.
-
Genome-Wide DNA Methylation and Transcriptome Analyses Reveal Epigenetic and Genetic Mechanisms Underlying Sex Maintenance of Adult Chinese Alligator.Front Genet. 2021 Mar 11;12:655900. doi: 10.3389/fgene.2021.655900. eCollection 2021. Front Genet. 2021. PMID: 33777112 Free PMC article.
-
The role of DNA methylation in human trophoblast differentiation.Epigenetics. 2018;13(12):1154-1173. doi: 10.1080/15592294.2018.1549462. Epub 2018 Dec 5. Epigenetics. 2018. PMID: 30475094 Free PMC article.
-
Novel genetic and epigenetic factors of importance for inter-individual differences in drug disposition, response and toxicity.Pharmacol Ther. 2019 May;197:122-152. doi: 10.1016/j.pharmthera.2019.01.002. Epub 2019 Jan 22. Pharmacol Ther. 2019. PMID: 30677473 Free PMC article. Review.
-
Fail-tests of DNA methylation clocks, and development of a noise barometer for measuring epigenetic pressure of aging and disease.Aging (Albany NY). 2023 Sep 12;15(17):8552-8575. doi: 10.18632/aging.205046. Epub 2023 Sep 12. Aging (Albany NY). 2023. PMID: 37702598 Free PMC article.
References
-
- Berkyurek AC, Suetake I, Arita K, Takeshita K, Nakagawa A, Shirakawa M, et al. The DNA methyltransferase Dnmt1 directly interacts with the SET and RING finger-associated (SRA) domain of the multifunctional protein Uhrf1 to facilitate accession of the catalytic center to hemi-methylated DNA. J Biol Chem. 2014;289:379–386. doi: 10.1074/jbc.M113.523209. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

