Site-directed mutagenesis, in vivo electroporation and mass spectrometry in search for determinants of the subcellular targeting of Rab7b paralogue in the model eukaryote Paramecium octaurelia
- PMID: 27349314
- PMCID: PMC4933825
- DOI: 10.4081/ejh.2016.2612
Site-directed mutagenesis, in vivo electroporation and mass spectrometry in search for determinants of the subcellular targeting of Rab7b paralogue in the model eukaryote Paramecium octaurelia
Abstract
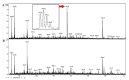
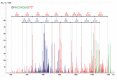
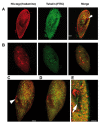
Protein products of the paralogous genes resulting from the whole genome duplication may acquire new function. The role of post-translational modifications (PTM) in proper targeting of Paramecium Rab7b paralogue - distinct from that of Rab7a directly involved in phagocytosis - was studied using point mutagenesis, proteomic analysis and double immunofluorescence after in vivo electroporation of the mutagenized protein. Here we show that substitution of Thr200 by Ala200 resulted in diminished incorporation of [P32] by 37.4% and of 32 [C14-]UDP-glucose by 24%, respectively, into recombinant Rab7b_200 in comparison to the non-mutagenized control. Double confocal imaging revealed that Rab7b_200 was mistargeted upon electroporation into living cells contrary to non- mutagenized recombinant Rab7b correctly incorporated in the cytostome area. We identified the peptide ion at m/z=677.63+ characteristic for the glycan group attached to Thr200 in Rab7b using nano LC-MS/MS and comparing the peptide map of this protein with that after deglycosylation with the mixture of five enzymes of different specificity. Based on the mass of this peptide ion and quantitative radioactive assays with [P32]and [C14-]UDP- glucose, the suggested composition of the adduct attached to Thr200 might be (Hex)1(HexNAc)1(Phos)3 or (HexNAc)1 (Deoxyhexose)1 (Phos)1 (HexA)1. These data indicate that PTM of Thr200 located in the hypervariable C-region of Rab7b in Paramecium is crucial for the proper localization/function of this protein. Moreover, these proteins differ also in other PTM: the number of phosphorylated amino acids in Rab7b is much higher than in Rab7a.
Conflict of interest statement
Conflict of interest: the authors report no conflics of interest.
Figures




Similar articles
-
Distinct expression, localization and function of two Rab7 proteins encoded by paralogous genes in a free-living model eukaryote.Acta Biochim Pol. 2011;58(4):597-607. Epub 2011 Oct 27. Acta Biochim Pol. 2011. PMID: 22030555
-
Cloning of two genes encoding Rab7 in Paramecium.Acta Biochim Pol. 2006;53(1):149-56. Epub 2005 Dec 19. Acta Biochim Pol. 2006. PMID: 16365637
-
Dynamics of Rab7b-dependent transport of sorting receptors.Traffic. 2012 Sep;13(9):1273-85. doi: 10.1111/j.1600-0854.2012.01388.x. Epub 2012 Jul 17. Traffic. 2012. PMID: 22708738
-
Early stages of functional diversification in the Rab GTPase gene family revealed by genomic and localization studies in Paramecium species.Mol Biol Cell. 2017 Apr 15;28(8):1101-1110. doi: 10.1091/mbc.E16-06-0361. Epub 2017 Mar 1. Mol Biol Cell. 2017. PMID: 28251922 Free PMC article.
-
Evolutionary conservancy of the endocytic and trafficking machinery in the unicellular eukaryote Paramecium.Biol Cell. 2003 Mar-Apr;95(2):69-74. doi: 10.1016/s0248-4900(03)00012-1. Biol Cell. 2003. PMID: 12799062 Review.
Cited by
-
Phosphorylation of Rab GTPases in the regulation of membrane trafficking.Traffic. 2020 Nov;21(11):712-719. doi: 10.1111/tra.12765. Epub 2020 Oct 19. Traffic. 2020. PMID: 32969543 Free PMC article.
-
The Rab7 subfamily across Paramecium aurelia species; evidence of high conservation in sequence and function.Small GTPases. 2020 Nov;11(6):421-429. doi: 10.1080/21541248.2018.1502056. Epub 2018 Aug 29. Small GTPases. 2020. PMID: 30156960 Free PMC article.
References
-
- Osińska M, Wiejak J, Wypych E, Bilski H, Bartosiewicz R, Wyroba E. Distinct expression, localization and function of two Rab7 proteins encoded by paralogous genes in a free-living model eukaryote. Acta Biochim Pol 2011;58:597-607. - PubMed
-
- Aury J-M, Jaillon O, Duret L, Noel B, Jubin C, Porcel B, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 2006;444:171-8. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

