Molecular Dynamics Driven Design of pH-Stabilized Mutants of MNEI, a Sweet Protein
- PMID: 27340829
- PMCID: PMC4920389
- DOI: 10.1371/journal.pone.0158372
Molecular Dynamics Driven Design of pH-Stabilized Mutants of MNEI, a Sweet Protein
Abstract
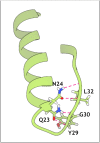
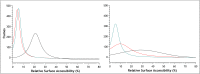
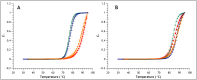
MNEI is a single chain derivative of monellin, a plant protein that can interact with the human sweet taste receptor, being therefore perceived as sweet. This unusual physiological activity makes MNEI a potential template for the design of new sugar replacers for the food and beverage industry. Unfortunately, applications of MNEI have been so far limited by its intrinsic sensitivity to some pH and temperature conditions, which could occur in industrial processes. Changes in physical parameters can, in fact, lead to irreversible protein denaturation, as well as aggregation and precipitation. It has been previously shown that the correlation between pH and stability in MNEI derives from the presence of a single glutamic residue in a hydrophobic pocket of the protein. We have used molecular dynamics to study the consequences, at the atomic level, of the protonation state of such residue and have identified the network of intramolecular interactions responsible for MNEI stability at acidic pH. Based on this information, we have designed a pH-independent, stabilized mutant of MNEI and confirmed its increased stability by both molecular modeling and experimental techniques.
Conflict of interest statement
Figures

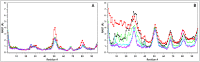
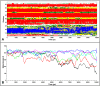
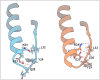
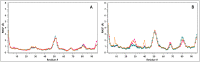
Similar articles
-
pH driven fibrillar aggregation of the super-sweet protein Y65R-MNEI: A step-by-step structural analysis.Biochim Biophys Acta Gen Subj. 2018 Apr;1862(4):808-815. doi: 10.1016/j.bbagen.2017.12.012. Epub 2017 Dec 27. Biochim Biophys Acta Gen Subj. 2018. PMID: 29288772
-
Design of sweet protein based sweeteners: hints from structure-function relationships.Food Chem. 2015 Apr 15;173:1179-86. doi: 10.1016/j.foodchem.2014.10.151. Epub 2014 Nov 6. Food Chem. 2015. PMID: 25466141
-
The Flexible Loop is a New Sweetness Determinant Site of the Sweet-Tasting Protein: Characterization of Novel Sweeter Mutants of the Single-Chain Monellin (MNEI).Chem Senses. 2019 Oct 17;44(8):607-614. doi: 10.1093/chemse/bjz057. Chem Senses. 2019. PMID: 31504288
-
Recent developments in the characterization and biotechnological production of sweet-tasting proteins.Appl Microbiol Biotechnol. 2000 Feb;53(2):145-51. doi: 10.1007/s002530050001. Appl Microbiol Biotechnol. 2000. PMID: 10709975 Review.
-
Bioprospecting and biotechnological insights into sweet-tasting proteins by microbial hosts-a review.Bioengineered. 2022 Apr;13(4):9815-9828. doi: 10.1080/21655979.2022.2061147. Bioengineered. 2022. PMID: 35435127 Free PMC article. Review.
Cited by
-
Enhanced Thermostability of Glucose Oxidase through Computer-Aided Molecular Design.Int J Mol Sci. 2018 Jan 31;19(2):425. doi: 10.3390/ijms19020425. Int J Mol Sci. 2018. PMID: 29385094 Free PMC article.
-
Insights from molecular dynamics simulations for computational protein design.Mol Syst Des Eng. 2017 Feb 1;2(1):9-33. doi: 10.1039/C6ME00083E. Epub 2017 Jan 9. Mol Syst Des Eng. 2017. PMID: 28239489 Free PMC article.
-
Molecular dynamics-derived rotamer libraries for d-amino acids within homochiral and heterochiral polypeptides.Protein Eng Des Sel. 2018 Jun 1;31(6):191-204. doi: 10.1093/protein/gzy016. Protein Eng Des Sel. 2018. PMID: 29992252 Free PMC article.
-
Expression of a single-chain monellin (MNEI) mutant with enhanced stability in transgenic mice milk.Transgenic Res. 2024 Aug;33(4):211-218. doi: 10.1007/s11248-024-00389-7. Epub 2024 Jun 11. Transgenic Res. 2024. PMID: 38858256
-
Metabolic Effects of the Sweet Protein MNEI as a Sweetener in Drinking Water. A Pilot Study of a High Fat Dietary Regimen in a Rodent Model.Nutrients. 2019 Nov 4;11(11):2643. doi: 10.3390/nu11112643. Nutrients. 2019. PMID: 31689911 Free PMC article.
References
-
- Morris JA, Cagan RH. Purification of monellin, the sweet principle of Dioscoreophyllum cumminsii. Biochim Biophys Acta. 1972;261: 114–122. - PubMed
-
- van der Wel H, Loeve K. Isolation and characterization of thaumatin I and II, the sweet-tasting proteins from Thaumatococcus daniellii Benth. Eur J Biochem. 1972;31: 221–225. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

