Aurora-C Interactions with Survivin and INCENP Reveal Shared and Distinct Features Compared with Aurora-B Chromosome Passenger Protein Complex
- PMID: 27332895
- PMCID: PMC4917241
- DOI: 10.1371/journal.pone.0157305
Aurora-C Interactions with Survivin and INCENP Reveal Shared and Distinct Features Compared with Aurora-B Chromosome Passenger Protein Complex
Abstract
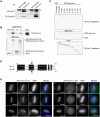
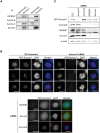
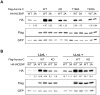
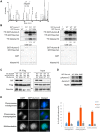
Aurora-C, a member of the Aurora kinase family that can complement Aurora-B function in mitosis is either moderately expressed or repressed in most adult somatic tissues but is active in early embryonic development and expressed at elevated levels in multiple human cancers. Aurora-C overexpression reportedly plays a role in tumorigenic transformation. We performed detailed characterization of Aurora-C interactions with members of the Chromosome Passenger Complex (CPC), Survivin and Inner Centromere Protein (INCENP) in reference to known Aurora-B interactions to understand the functional significance of Aurora-C overexpression in human cancer cells. The results revealed that silencing of Aurora-C or -B individually does not affect localization of the other kinase and the two kinases exist predominantly in independent complexes in vivo. Presence of Aurora-C and -B in molecular complexes of varying as well as overlapping sizes and co-existence in INCENP overexpressing cells indicated oligomerization of ternary complexes under different physiological conditions in vivo. Furthermore, Aurora-C and -B stabilized INCENP through interaction with and phosphorylation of the IN box domain while Aurora-C was activated following Survivin phosphorylation on Serine 20. Phosphorylation of Survivin residue Serine 20 by Aurora-C and -B appears important for proper chromosome segregation. Taken together, our study suggests that Aurora-C, expressed at low levels in somatic cells, functions as a catalytic component of the CPC together with Aurora-B through mitosis. Elevated expression of Aurora-C in cancer cells alters the structural and functional characteristics of the Aurora-B-CPC leading to chromosomal instability.
Conflict of interest statement
Figures


Similar articles
-
Targeting the INCENP IN-box-Aurora B interaction to inhibit CPC activity in vivo.Open Biol. 2014 Nov;4(11):140163. doi: 10.1098/rsob.140163. Open Biol. 2014. PMID: 25392451 Free PMC article.
-
Overexpression of Aurora-C interferes with the spindle checkpoint by promoting the degradation of Aurora-B.Cell Death Dis. 2014 Mar 6;5(3):e1106. doi: 10.1038/cddis.2014.37. Cell Death Dis. 2014. PMID: 24603334 Free PMC article.
-
Aurora-B phosphorylation in vitro identifies a residue of survivin that is essential for its localization and binding to inner centromere protein (INCENP) in vivo.J Biol Chem. 2004 Feb 13;279(7):5655-60. doi: 10.1074/jbc.M311299200. Epub 2003 Nov 10. J Biol Chem. 2004. PMID: 14610074
-
Role of chromosomal passenger complex in chromosome segregation and cytokinesis.Cell Struct Funct. 2001 Dec;26(6):653-7. doi: 10.1247/csf.26.653. Cell Struct Funct. 2001. PMID: 11942622 Review.
-
Chromosomal passengers and the (aurora) ABCs of mitosis.Trends Cell Biol. 2001 Feb;11(2):49-54. doi: 10.1016/s0962-8924(00)01880-8. Trends Cell Biol. 2001. PMID: 11166196 Review.
Cited by
-
An Aurora kinase inhibitor, AMG900, inhibits glioblastoma cell proliferation by disrupting mitotic progression.Cancer Med. 2018 Nov;7(11):5589-5603. doi: 10.1002/cam4.1771. Epub 2018 Sep 17. Cancer Med. 2018. PMID: 30221846 Free PMC article.
-
BIRC5/Survivin is a novel ATG12-ATG5 conjugate interactor and an autophagy-induced DNA damage suppressor in human cancer and mouse embryonic fibroblast cells.Autophagy. 2020 Jul;16(7):1296-1313. doi: 10.1080/15548627.2019.1671643. Epub 2019 Oct 15. Autophagy. 2020. PMID: 31612776 Free PMC article.
-
The Relevance of Aurora Kinase Inhibition in Hematological Malignancies.Cancer Diagn Progn. 2021 Jul 3;1(3):111-126. doi: 10.21873/cdp.10016. eCollection 2021 Jul-Aug. Cancer Diagn Progn. 2021. PMID: 35399305 Free PMC article. Review.
-
Genetic Interactions between the Aurora Kinases Reveal New Requirements for AURKB and AURKC during Oocyte Meiosis.Curr Biol. 2018 Nov 5;28(21):3458-3468.e5. doi: 10.1016/j.cub.2018.08.052. Epub 2018 Oct 25. Curr Biol. 2018. PMID: 30415701 Free PMC article.
-
Aurora kinases: novel therapy targets in cancers.Oncotarget. 2017 Apr 4;8(14):23937-23954. doi: 10.18632/oncotarget.14893. Oncotarget. 2017. PMID: 28147341 Free PMC article. Review.
References
-
- Carmena M, Earnshaw WC. The cellular geography of aurora kinases. Nat Rev Mol Cell Biol. 2003;4(11):842–54. - PubMed
-
- Jeyaprakash AA, Klein UR, Lindner D, Ebert J, Nigg EA, Conti E. Structure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell. 2007;131(2):271–85. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

