Membrane-Spanning Sequences in Endoplasmic Reticulum Proteins Promote Phospholipid Flip-Flop
- PMID: 27332127
- PMCID: PMC4919424
- DOI: 10.1016/j.bpj.2016.05.023
Membrane-Spanning Sequences in Endoplasmic Reticulum Proteins Promote Phospholipid Flip-Flop
Abstract
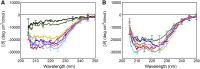
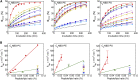
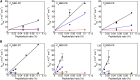
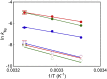
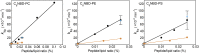
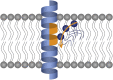
The mechanism whereby phospholipids rapidly flip-flop in the endoplasmic reticulum (ER) membrane remains unknown. We previously demonstrated that the presence of a hydrophilic residue in the center of the model transmembrane peptide sequence effectively promoted phospholipid flip-flop and that hydrophilic residues composed 4.5% of the central regions of the membrane-spanning sequences of human ER membrane proteins predicted by SOSUI software. We hypothesized that ER proteins with hydrophilic residues might play a critical role in promoting flip-flop. Here, we evaluated the flip rate of fluorescently labeled lipids in vesicles containing each of the 11 synthetic peptides of membrane-spanning sequences, using a dithionite-quenching assay. Although the flippase activities of nine peptides were unexpectedly low, the peptides based on the EDEM1 and SPAST proteins showed enhanced flippase activity with three different fluorescently labeled lipids. The substitution of hydrophobic Ala with His or Arg in the central region of the EDEM1 or SPAST peptides, respectively, attenuated their ability to flip phospholipids. Interestingly, substituting Ala with Arg or His at a location outside of the central region of EDEM1 or SPAST, respectively, also affected the enhancement of flip-flop. These results indicated that both Arg and His are important for the ability of these two peptides to increase the flip rates. The EDEM1 peptide exhibited high activity at significantly low peptide concentrations, suggesting that the same side positioning of Arg and His in α-helix structure is critical for the flip-flop promotion and that the EDEM1 protein is a candidate flippase in the ER.
Copyright © 2016 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Phospholipid flop induced by transmembrane peptides in model membranes is modulated by lipid composition.Biochemistry. 2003 Jan 14;42(1):231-7. doi: 10.1021/bi0268403. Biochemistry. 2003. PMID: 12515559
-
Flip-flop of fluorescently labeled phospholipids in proteoliposomes reconstituted with Saccharomyces cerevisiae microsomal proteins.Eukaryot Cell. 2007 Sep;6(9):1625-34. doi: 10.1128/EC.00198-07. Epub 2007 Jul 6. Eukaryot Cell. 2007. PMID: 17616631 Free PMC article.
-
Effect of lipid composition and amino acid sequence upon transmembrane peptide-accelerated lipid transleaflet diffusion (flip-flop).Biochim Biophys Acta. 2016 Aug;1858(8):1812-20. doi: 10.1016/j.bbamem.2016.04.011. Epub 2016 Apr 27. Biochim Biophys Acta. 2016. PMID: 27131444
-
Phospholipid flip-flop in biogenic membranes: what is needed to connect opposite sides.Semin Cell Dev Biol. 2002 Jun;13(3):163-70. doi: 10.1016/s1084-9521(02)00044-7. Semin Cell Dev Biol. 2002. PMID: 12137736 Review.
-
The mystery of phospholipid flip-flop in biogenic membranes.Cell Mol Biol Lett. 2005;10(1):101-21. Cell Mol Biol Lett. 2005. PMID: 15809683 Review.
Cited by
-
Genome-wide CRISPR screen reveals CLPTM1L as a lipid scramblase required for efficient glycosylphosphatidylinositol biosynthesis.Proc Natl Acad Sci U S A. 2022 Apr 5;119(14):e2115083119. doi: 10.1073/pnas.2115083119. Epub 2022 Mar 28. Proc Natl Acad Sci U S A. 2022. PMID: 35344438 Free PMC article.
-
Phospholipid Scrambling by G Protein-Coupled Receptors.Annu Rev Biophys. 2022 May 9;51:39-61. doi: 10.1146/annurev-biophys-090821-083030. Epub 2021 Dec 21. Annu Rev Biophys. 2022. PMID: 34932914 Free PMC article. Review.
-
Lipid Droplets in the Pathogenesis of Hereditary Spastic Paraplegia.Front Mol Biosci. 2021 May 10;8:673977. doi: 10.3389/fmolb.2021.673977. eCollection 2021. Front Mol Biosci. 2021. PMID: 34041268 Free PMC article. Review.
-
Development of membrane-insertable lipid scrambling peptides: A time-resolved small-angle neutron scattering study.Struct Dyn. 2021 Mar 19;8(2):024301. doi: 10.1063/4.0000045. eCollection 2021 Mar. Struct Dyn. 2021. PMID: 33758768 Free PMC article.
-
Microtubule-dependent and independent roles of spastin in lipid droplet dispersion and biogenesis.Life Sci Alliance. 2020 Apr 22;3(6):e202000715. doi: 10.26508/lsa.202000715. Print 2020 Jun. Life Sci Alliance. 2020. PMID: 32321733 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

