Transcription factor Bcl11b sustains iNKT1 and iNKT2 cell programs, restricts iNKT17 cell program, and governs iNKT cell survival
- PMID: 27330109
- PMCID: PMC4941433
- DOI: 10.1073/pnas.1521846113
Transcription factor Bcl11b sustains iNKT1 and iNKT2 cell programs, restricts iNKT17 cell program, and governs iNKT cell survival
Abstract
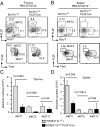
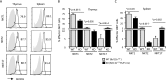
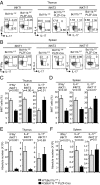
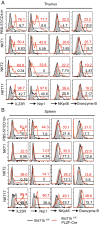
Invariant natural killer T (iNKT) cells are innate-like T cells that recognize glycolipid antigens and play critical roles in regulation of immune responses. Based on expression of the transcription factors (TFs) Tbet, Plzf, and Rorγt, iNKT cells have been classified in effector subsets that emerge in the thymus, namely, iNKT1, iNKT2, and iNKT17. Deficiency in the TF Bcl11b in double-positive (DP) thymocytes has been shown to cause absence of iNKT cells in the thymus and periphery due to defective self glycolipid processing and presentation by DP thymocytes and undefined intrinsic alterations in iNKT precursors. We used a model of cre-mediated postselection deletion of Bcl11b in iNKT cells to determine its intrinsic role in these cells. We found that Bcl11b is expressed equivalently in all three effector iNKT subsets, and its removal caused a reduction in the numbers of iNKT1 and iNKT2 cells, but not in the numbers of iNKT17 cells. Additionally, we show that Bcl11b sustains subset-specific cytokine production by iNKT1 and iNKT2 cells and restricts expression of iNKT17 genes in iNKT1 and iNKT2 subsets, overall restraining the iNKT17 program in iNKT cells. The total numbers of iNKT cells were reduced in the absence of Bcl11b both in the thymus and periphery, associated with the decrease in iNKT1 and iNKT2 cell numbers and decrease in survival, related to changes in survival/apoptosis genes. Thus, these results extend our understanding of the role of Bcl11b in iNKT cells beyond their selection and demonstrate that Bcl11b is a key regulator of iNKT effector subsets, their function, identity, and survival.
Keywords: iNKT cell program; iNKT1 effector cells; iNKT17 effector cells; iNKT2 effector cells; transcription factor Bcl11b.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Unraveling Natural Killer T-Cells Development.Front Immunol. 2018 Jan 9;8:1950. doi: 10.3389/fimmu.2017.01950. eCollection 2017. Front Immunol. 2018. PMID: 29375573 Free PMC article. Review.
-
β-Catenin is required for the differentiation of iNKT2 and iNKT17 cells that augment IL-25-dependent lung inflammation.BMC Immunol. 2015 Oct 19;16:62. doi: 10.1186/s12865-015-0121-0. BMC Immunol. 2015. PMID: 26482437 Free PMC article.
-
iNKT subsets differ in their developmental and functional requirements on Foxo1.Proc Natl Acad Sci U S A. 2021 Nov 16;118(46):e2105950118. doi: 10.1073/pnas.2105950118. Proc Natl Acad Sci U S A. 2021. PMID: 34772808 Free PMC article.
-
The differentiation of ROR-γt expressing iNKT17 cells is orchestrated by Runx1.Sci Rep. 2017 Aug 1;7(1):7018. doi: 10.1038/s41598-017-07365-8. Sci Rep. 2017. PMID: 28765611 Free PMC article.
-
mTOR and its tight regulation for iNKT cell development and effector function.Mol Immunol. 2015 Dec;68(2 Pt C):536-45. doi: 10.1016/j.molimm.2015.07.022. Epub 2015 Aug 4. Mol Immunol. 2015. PMID: 26253278 Free PMC article. Review.
Cited by
-
TET proteins regulate the lineage specification and TCR-mediated expansion of iNKT cells.Nat Immunol. 2017 Jan;18(1):45-53. doi: 10.1038/ni.3630. Epub 2016 Nov 21. Nat Immunol. 2017. PMID: 27869820 Free PMC article.
-
Myeloid/natural killer (NK) cell precursor acute leukemia as a distinct leukemia type.Sci Adv. 2023 Dec 15;9(50):eadj4407. doi: 10.1126/sciadv.adj4407. Epub 2023 Dec 13. Sci Adv. 2023. PMID: 38091391 Free PMC article.
-
Coexpression of YY1 Is Required to Elaborate the Effector Functions Controlled by PLZF in NKT Cells.J Immunol. 2019 Aug 1;203(3):627-638. doi: 10.4049/jimmunol.1900055. Epub 2019 Jun 21. J Immunol. 2019. PMID: 31227579 Free PMC article.
-
Bcl11b is essential for licensing Th2 differentiation during helminth infection and allergic asthma.Nat Commun. 2018 Apr 26;9(1):1679. doi: 10.1038/s41467-018-04111-0. Nat Commun. 2018. PMID: 29700302 Free PMC article.
-
Unraveling Natural Killer T-Cells Development.Front Immunol. 2018 Jan 9;8:1950. doi: 10.3389/fimmu.2017.01950. eCollection 2017. Front Immunol. 2018. PMID: 29375573 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

