Molecular pathological classification of colorectal cancer
- PMID: 27325016
- PMCID: PMC4978761
- DOI: 10.1007/s00428-016-1956-3
Molecular pathological classification of colorectal cancer
Abstract
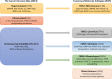
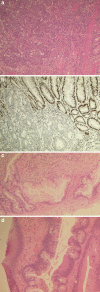
Colorectal cancer (CRC) shows variable underlying molecular changes with two major mechanisms of genetic instability: chromosomal instability and microsatellite instability. This review aims to delineate the different pathways of colorectal carcinogenesis and provide an overview of the most recent advances in molecular pathological classification systems for colorectal cancer. Two molecular pathological classification systems for CRC have recently been proposed. Integrated molecular analysis by The Cancer Genome Atlas project is based on a wide-ranging genomic and transcriptomic characterisation study of CRC using array-based and sequencing technologies. This approach classified CRC into two major groups consistent with previous classification systems: (1) ∼16 % hypermutated cancers with either microsatellite instability (MSI) due to defective mismatch repair (∼13 %) or ultramutated cancers with DNA polymerase epsilon proofreading mutations (∼3 %); and (2) ∼84 % non-hypermutated, microsatellite stable (MSS) cancers with a high frequency of DNA somatic copy number alterations, which showed common mutations in APC, TP53, KRAS, SMAD4, and PIK3CA. The recent Consensus Molecular Subtypes (CMS) Consortium analysing CRC expression profiling data from multiple studies described four CMS groups: almost all hypermutated MSI cancers fell into the first category CMS1 (MSI-immune, 14 %) with the remaining MSS cancers subcategorised into three groups of CMS2 (canonical, 37 %), CMS3 (metabolic, 13 %) and CMS4 (mesenchymal, 23 %), with a residual unclassified group (mixed features, 13 %). Although further research is required to validate these two systems, they may be useful for clinical trial designs and future post-surgical adjuvant treatment decisions, particularly for tumours with aggressive features or predicted responsiveness to immune checkpoint blockade.
Keywords: Cancer; Chromosomal instability; Colorectal; Consensus molecular subtypes; Defective mismatch repair; Hypermutant; Microsatellite instability; Mutation; Polymerase epsilon; Serrated pathway; Somatic copy number alterations; The Cancer Genome Atlas; Ultramutant.
Figures
Similar articles
-
Consensus molecular subtype classification of colorectal adenomas.J Pathol. 2018 Nov;246(3):266-276. doi: 10.1002/path.5129. Epub 2018 Aug 31. J Pathol. 2018. PMID: 29968252 Free PMC article.
-
Genomic and transcriptomic characterization of heterogeneous immune subgroups of microsatellite instability-high colorectal cancers.J Immunother Cancer. 2021 Dec;9(12):e003414. doi: 10.1136/jitc-2021-003414. J Immunother Cancer. 2021. PMID: 34903553 Free PMC article.
-
[Characterization of patients with sporadic colorectal cancer following the new Consensus Molecular Subtypes (CMS)].Rev Med Chil. 2017 Apr;145(4):419-430. doi: 10.4067/S0034-98872017000400001. Rev Med Chil. 2017. PMID: 28748988 Spanish.
-
Genetics and Genetic Biomarkers in Sporadic Colorectal Cancer.Gastroenterology. 2015 Oct;149(5):1177-1190.e3. doi: 10.1053/j.gastro.2015.06.047. Epub 2015 Jul 26. Gastroenterology. 2015. PMID: 26216840 Free PMC article. Review.
-
Is There a Role for Programmed Death Ligand-1 Testing and Immunotherapy in Colorectal Cancer With Microsatellite Instability? Part I-Colorectal Cancer: Microsatellite Instability, Testing, and Clinical Implications.Arch Pathol Lab Med. 2018 Jan;142(1):17-25. doi: 10.5858/arpa.2017-0040-RA. Epub 2017 Nov 16. Arch Pathol Lab Med. 2018. PMID: 29144791 Review.
Cited by
-
Aloperine Inhibits Proliferation and Promotes Apoptosis in Colorectal Cancer Cells by Regulating the circNSUN2/miR-296-5p/STAT3 Pathway.Drug Des Devel Ther. 2021 Feb 25;15:857-870. doi: 10.2147/DDDT.S288473. eCollection 2021. Drug Des Devel Ther. 2021. PMID: 33664565 Free PMC article.
-
Left-sided colorectal cancer distinct in indigenous African patients compared to other ethnic groups in South Africa.BMC Cancer. 2022 Oct 24;22(1):1089. doi: 10.1186/s12885-022-10185-3. BMC Cancer. 2022. PMID: 36280820 Free PMC article.
-
Construction of a long noncoding RNA-based competing endogenous RNA network and prognostic signatures of left- and right-side colon cancer.Cancer Cell Int. 2021 Apr 15;21(1):211. doi: 10.1186/s12935-021-01901-3. Cancer Cell Int. 2021. PMID: 33858429 Free PMC article.
-
Evolutionary history of adenomas to colorectal cancer in FAP families.Front Genet. 2024 Jul 3;15:1391851. doi: 10.3389/fgene.2024.1391851. eCollection 2024. Front Genet. 2024. PMID: 39021676 Free PMC article.
-
The clinical and prognostic evaluation of GRP94 immunoexpression in Caucasian patients with colorectal adenocarcinoma.Prz Gastroenterol. 2019;14(2):140-147. doi: 10.5114/pg.2019.85898. Epub 2019 Jul 5. Prz Gastroenterol. 2019. PMID: 31616529 Free PMC article.
References
-
- Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin D, Forman D, Bray F (2013) GLOBOCAN 2012 v1.0, cancer incidence and mortality worldwide: IARC CancerBase No. 11 [Internet]. International Agency for Research on Cancer. http://globocan.iarc.fr. Accessed 11/03/2016
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

