Chloroplast DNA Structural Variation, Phylogeny, and Age of Divergence among Diploid Cotton Species
- PMID: 27309527
- PMCID: PMC4911064
- DOI: 10.1371/journal.pone.0157183
Chloroplast DNA Structural Variation, Phylogeny, and Age of Divergence among Diploid Cotton Species
Abstract
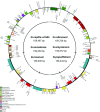
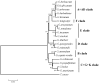
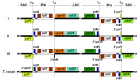
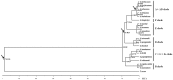
The cotton genus (Gossypium spp.) contains 8 monophyletic diploid genome groups (A, B, C, D, E, F, G, K) and a single allotetraploid clade (AD). To gain insight into the phylogeny of Gossypium and molecular evolution of the chloroplast genome in this group, we performed a comparative analysis of 19 Gossypium chloroplast genomes, six reported here for the first time. Nucleotide distance in non-coding regions was about three times that of coding regions. As expected, distances were smaller within than among genome groups. Phylogenetic topologies based on nucleotide and indel data support for the resolution of the 8 genome groups into 6 clades. Phylogenetic analysis of indel distribution among the 19 genomes demonstrates contrasting evolutionary dynamics in different clades, with a parallel genome downsizing in two genome groups and a biased accumulation of insertions in the clade containing the cultivated cottons leading to large (for Gossypium) chloroplast genomes. Divergence time estimates derived from the cpDNA sequence suggest that the major diploid clades had diverged approximately 10 to 11 million years ago. The complete nucleotide sequences of 6 cpDNA genomes are provided, offering a resource for cytonuclear studies in Gossypium.
Conflict of interest statement
Figures

Similar articles
-
Molecular evolution of the plastid genome during diversification of the cotton genus.Mol Phylogenet Evol. 2017 Jul;112:268-276. doi: 10.1016/j.ympev.2017.04.014. Epub 2017 Apr 13. Mol Phylogenet Evol. 2017. PMID: 28414099
-
Analysis of complete nucleotide sequences of 12 Gossypium chloroplast genomes: origin and evolution of allotetraploids.PLoS One. 2012;7(8):e37128. doi: 10.1371/journal.pone.0037128. Epub 2012 Aug 2. PLoS One. 2012. PMID: 22876273 Free PMC article.
-
Rapid evolutionary divergence of diploid and allotetraploid Gossypium mitochondrial genomes.BMC Genomics. 2017 Nov 13;18(1):876. doi: 10.1186/s12864-017-4282-5. BMC Genomics. 2017. PMID: 29132310 Free PMC article.
-
Intergenomic gene transfer in diploid and allopolyploid Gossypium.BMC Plant Biol. 2019 Nov 12;19(1):492. doi: 10.1186/s12870-019-2041-2. BMC Plant Biol. 2019. PMID: 31718541 Free PMC article.
-
Sequencing Multiple Cotton Genomes Reveals Complex Structures and Lays Foundation for Breeding.Front Plant Sci. 2020 Sep 16;11:560096. doi: 10.3389/fpls.2020.560096. eCollection 2020. Front Plant Sci. 2020. PMID: 33042184 Free PMC article. Review.
Cited by
-
Taxonomical Evaluation of Plant Chloroplastic Markers by Bayesian Classifier.Front Plant Sci. 2022 Feb 3;12:782663. doi: 10.3389/fpls.2021.782663. eCollection 2021. Front Plant Sci. 2022. PMID: 35185949 Free PMC article.
-
Genome-Wide Identification and Functional Investigation of 1-Aminocyclopropane-1-carboxylic Acid Oxidase (ACO) Genes in Cotton.Plants (Basel). 2021 Aug 18;10(8):1699. doi: 10.3390/plants10081699. Plants (Basel). 2021. PMID: 34451744 Free PMC article.
-
Genome-wide analysis of zinc finger-homeodomain (ZF-HD) transcription factors in diploid and tetraploid cotton.Funct Integr Genomics. 2022 Dec;22(6):1269-1281. doi: 10.1007/s10142-022-00913-0. Epub 2022 Nov 12. Funct Integr Genomics. 2022. PMID: 36369302
-
Insights into the Evolution of the New World Diploid Cottons (Gossypium, Subgenus Houzingenia) Based on Genome Sequencing.Genome Biol Evol. 2019 Jan 1;11(1):53-71. doi: 10.1093/gbe/evy256. Genome Biol Evol. 2019. PMID: 30476109 Free PMC article.
-
Nucleotide Evolution, Domestication Selection, and Genetic Relationships of Chloroplast Genomes in the Economically Important Crop Genus Gossypium.Front Plant Sci. 2022 Apr 15;13:873788. doi: 10.3389/fpls.2022.873788. eCollection 2022. Front Plant Sci. 2022. PMID: 35498673 Free PMC article.
References
-
- Wendel JF, Cronn RC. Polyploidy and the evolutionary history of cotton. Adv Agron. 2003;78:139–186. 10.1016/s0065-2113(02)78004-8 - DOI
-
- Wendel JF, Grover CE. Taxonomy and evolution of the cotton genus In: Fang D and Percy R, editors. Cotton, Agronomy. Madison, WI: Monograph 24, ASA-CSSA-SSSA; 2015. in press.
-
- Wendel J, Brubaker C, Seelanan T. The origin and evolution of Gossypium In: Physiology of Cotton. Edited by Stewart J, Oosterhuis D, Heitholt J, Mauney J: Springer; Netherlands: 2010;1–18.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

