Phosphatase ABI1 and okadaic acid-sensitive phosphoprotein phosphatases inhibit salt stress-activated SnRK2.4 kinase
- PMID: 27297076
- PMCID: PMC4907068
- DOI: 10.1186/s12870-016-0817-1
Phosphatase ABI1 and okadaic acid-sensitive phosphoprotein phosphatases inhibit salt stress-activated SnRK2.4 kinase
Abstract
Background: SNF1-related protein kinases 2 (SnRK2s) are key regulators of the plant response to osmotic stress. They are transiently activated in response to drought and salinity. Based on a phylogenetic analysis SnRK2s are divided into three groups. The classification correlates with their response to abscisic acid (ABA); group 1 consists SnRK2s non-activated in response to ABA, group 2, kinases non-activated or weakly activated (depending on the plant species) by ABA treatment, and group 3, ABA-activated kinases. The activity of all SnRK2s is regulated by phosphorylation. It is well established that clade A phosphoprotein phosphatases 2C (PP2Cs) are negative regulators of ABA-activated SnRK2s, whereas regulators of SnRK2s from group 1 remain unidentified.
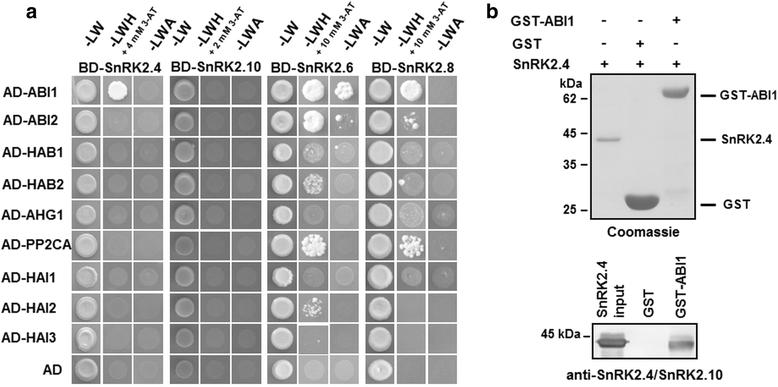
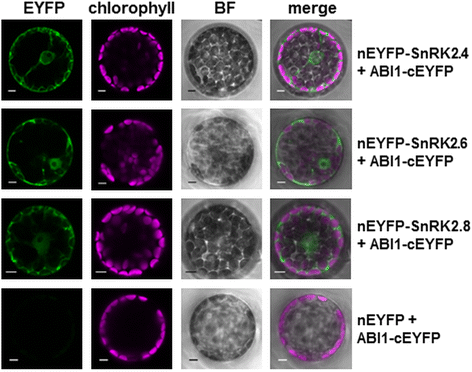
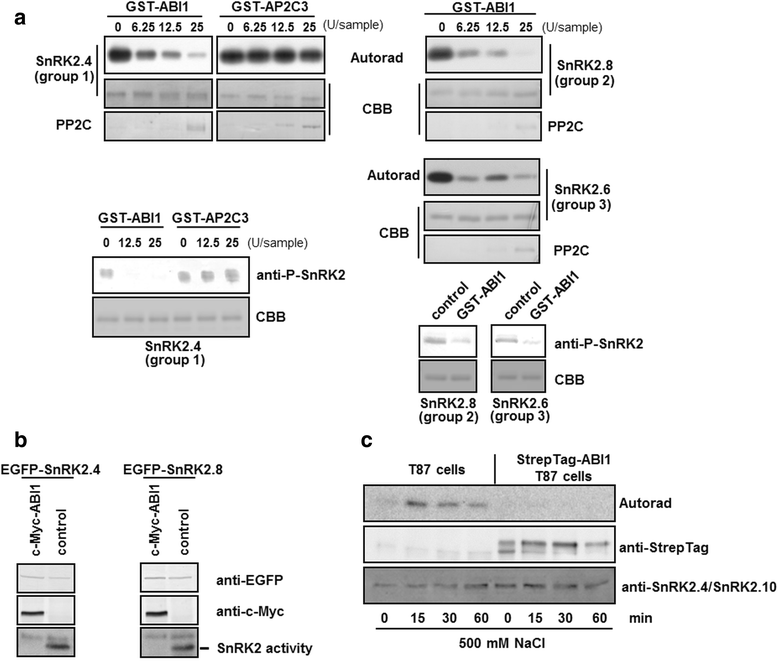
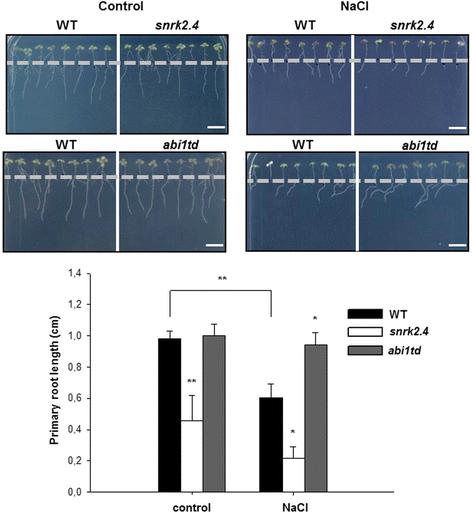
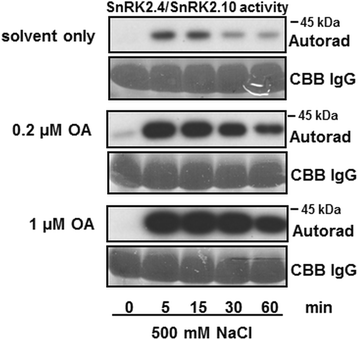
Results: Here, we show that ABI1, a PP2C clade A phosphatase, interacts with SnRK2.4, member of group 1 of the SnRK2 family, dephosphorylates Ser158, whose phosphorylation is needed for the kinase activity, and inhibits the kinase, both in vitro and in vivo. Our data indicate that ABI1 and the kinase regulate primary root growth in response to salinity; the phenotype of ABI1 knockout mutant (abi1td) exposed to salt stress is opposite to that of the snrk2.4 mutant. Moreover, we show that the activity of SnRK2s from group 1 is additionally regulated by okadaic acid-sensitive phosphatase(s) from the phosphoprotein phosphatase (PPP) family.
Conclusions: Phosphatase ABI1 and okadaic acid-sensitive phosphatases of the PPP family are negative regulators of salt stress-activated SnRK2.4. The results show that ABI1 inhibits not only the ABA-activated SnRK2s but also at least one ABA-non-activated SnRK2, suggesting that the phosphatase is involved in the cross talk between ABA-dependent and ABA-independent stress signaling pathways in plants.
Keywords: ABI1; Arabidopsis thaliana; Osmotic stress signaling; PP2C; PPP; Phosphoprotein phosphatases; SNF1-related protein kinases 2; Salinity; SnRK2.
Figures
Similar articles
-
The Multifaceted Regulation of SnRK2 Kinases.Cells. 2021 Aug 24;10(9):2180. doi: 10.3390/cells10092180. Cells. 2021. PMID: 34571829 Free PMC article. Review.
-
Protein phosphatase type 2C PP2CA together with ABI1 inhibits SnRK2.4 activity and regulates plant responses to salinity.Plant Signal Behav. 2016 Dec;11(12):e1253647. doi: 10.1080/15592324.2016.1253647. Plant Signal Behav. 2016. PMID: 27901636 Free PMC article.
-
Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis.Plant Cell Physiol. 2009 Dec;50(12):2123-32. doi: 10.1093/pcp/pcp147. Plant Cell Physiol. 2009. PMID: 19880399
-
Osmotic signaling releases PP2C-mediated inhibition of Arabidopsis SnRK2s via the receptor-like cytoplasmic kinase BIK1.EMBO J. 2024 Dec;43(23):6076-6103. doi: 10.1038/s44318-024-00277-0. Epub 2024 Oct 21. EMBO J. 2024. PMID: 39433899 Free PMC article.
-
Regulation of Arabidopsis MAPKKK18 by ABI1 and SnRK2, components of the ABA signaling pathway.Plant Signal Behav. 2016;11(4):e1139277. doi: 10.1080/15592324.2016.1139277. Plant Signal Behav. 2016. PMID: 26852793 Free PMC article. Review.
Cited by
-
SnRK2 subfamily I protein kinases regulate ethylene biosynthesis by phosphorylating HB transcription factors to induce ACO1 expression in apple.New Phytol. 2022 May;234(4):1262-1277. doi: 10.1111/nph.18040. Epub 2022 Mar 22. New Phytol. 2022. PMID: 35182082 Free PMC article.
-
The Multifaceted Regulation of SnRK2 Kinases.Cells. 2021 Aug 24;10(9):2180. doi: 10.3390/cells10092180. Cells. 2021. PMID: 34571829 Free PMC article. Review.
-
Functional Study of BpPP2C1 Revealed Its Role in Salt Stress in Betula platyphylla.Front Plant Sci. 2021 Jan 14;11:617635. doi: 10.3389/fpls.2020.617635. eCollection 2020. Front Plant Sci. 2021. PMID: 33519877 Free PMC article.
-
Regulation of Plant Responses to Salt Stress.Int J Mol Sci. 2021 Apr 28;22(9):4609. doi: 10.3390/ijms22094609. Int J Mol Sci. 2021. PMID: 33924753 Free PMC article. Review.
-
Protein Phosphatases Type 2C Group A Interact with and Regulate the Stability of ACC Synthase 7 in Arabidopsis.Cells. 2020 Apr 15;9(4):978. doi: 10.3390/cells9040978. Cells. 2020. PMID: 32326656 Free PMC article.
References
-
- Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, et al. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009;50(7):1345–1363. doi: 10.1093/pcp/pcp083. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

