Wnt/β-catenin pathway transactivates microRNA-150 that promotes EMT of colorectal cancer cells by suppressing CREB signaling
- PMID: 27285761
- PMCID: PMC5173152
- DOI: 10.18632/oncotarget.9893
Wnt/β-catenin pathway transactivates microRNA-150 that promotes EMT of colorectal cancer cells by suppressing CREB signaling
Abstract
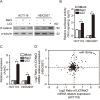
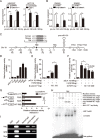
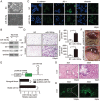
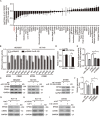
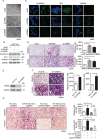
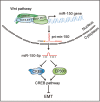
A hallmark of aberrant activation of the Wnt/β-catenin signaling pathway has been observed in most colorectal cancers (CRC), but little is known about the role of non-coding RNAs regulated by this pathway. Here, we found that miR-150 was the most significantly upregulated microRNA responsive to elevated of Wnt/β-catenin signaling activity in both HCT116 and HEK293T cells. Mechanistically, the β-catenin/LEF1 complex binds to the conserved TCF/LEF1-binding element in the miR-150 promoter and thereby transactivates its expression. Enforced expression of miR-150 in HCT116 cell line transformed cells into a spindle shape with higher migration and invasion activity. miR-150 markedly suppressed the CREB signaling pathway by targeting its core transcription factors CREB1 and EP300. Knockdown of CREB1 or EP300 and knockout of CREB1 by CRISPR/Cas9 phenocopied the epithelial-mesenchymal transition (EMT) observed in HCT116 cells in response to miR-150 overexpression. In summary, our data indicate that miR-150 is a novel Wnt effector that may significantly enhance EMT of CRC cells by targeting the CREB signaling pathway.
Keywords: CREB; EMT; Wnt/β-catenin; colorectal cancer; miR-150.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
OVOL2, an Inhibitor of WNT Signaling, Reduces Invasive Activities of Human and Mouse Cancer Cells and Is Down-regulated in Human Colorectal Tumors.Gastroenterology. 2016 Mar;150(3):659-671.e16. doi: 10.1053/j.gastro.2015.11.041. Epub 2015 Nov 24. Gastroenterology. 2016. PMID: 26619963
-
MiR-452 promotes an aggressive colorectal cancer phenotype by regulating a Wnt/β-catenin positive feedback loop.J Exp Clin Cancer Res. 2018 Sep 25;37(1):238. doi: 10.1186/s13046-018-0879-z. J Exp Clin Cancer Res. 2018. PMID: 30253791 Free PMC article.
-
IWR-1 inhibits epithelial-mesenchymal transition of colorectal cancer cells through suppressing Wnt/β-catenin signaling as well as survivin expression.Oncotarget. 2015 Sep 29;6(29):27146-59. doi: 10.18632/oncotarget.4354. Oncotarget. 2015. PMID: 26450645 Free PMC article.
-
Distinctive microRNA signature associated of neoplasms with the Wnt/β-catenin signaling pathway.Cell Signal. 2013 Dec;25(12):2805-11. doi: 10.1016/j.cellsig.2013.09.006. Epub 2013 Sep 13. Cell Signal. 2013. PMID: 24041653 Review.
-
Interplay between microRNAs and WNT/β-catenin signalling pathway regulates epithelial-mesenchymal transition in cancer.Eur J Cancer. 2015 Aug;51(12):1638-49. doi: 10.1016/j.ejca.2015.04.021. Epub 2015 May 26. Eur J Cancer. 2015. PMID: 26025765 Review.
Cited by
-
MicroRNA-150 suppresses triple-negative breast cancer metastasis through targeting HMGA2.Onco Targets Ther. 2018 Apr 24;11:2319-2332. doi: 10.2147/OTT.S161996. eCollection 2018. Onco Targets Ther. 2018. PMID: 29731640 Free PMC article.
-
Exosomes in metastasis of colorectal cancers: Friends or foes?World J Gastrointest Oncol. 2023 May 15;15(5):731-756. doi: 10.4251/wjgo.v15.i5.731. World J Gastrointest Oncol. 2023. PMID: 37275444 Free PMC article. Review.
-
MicroRNAs in the prognosis and therapy of colorectal cancer: From bench to bedside.World J Gastroenterol. 2018 Jul 21;24(27):2949-2973. doi: 10.3748/wjg.v24.i27.2949. World J Gastroenterol. 2018. PMID: 30038463 Free PMC article. Review.
-
Exosomal miR-128-3p Promotes Epithelial-to-Mesenchymal Transition in Colorectal Cancer Cells by Targeting FOXO4 via TGF-β/SMAD and JAK/STAT3 Signaling.Front Cell Dev Biol. 2021 Feb 9;9:568738. doi: 10.3389/fcell.2021.568738. eCollection 2021. Front Cell Dev Biol. 2021. Retraction in: Front Cell Dev Biol. 2024 Jul 19;12:1459522. doi: 10.3389/fcell.2024.1459522. PMID: 33634112 Free PMC article. Retracted.
-
FSH induces EMT in ovarian cancer via ALKBH5-regulated Snail m6A demethylation.Theranostics. 2024 Mar 3;14(5):2151-2166. doi: 10.7150/thno.94161. eCollection 2024. Theranostics. 2024. PMID: 38505602 Free PMC article.
References
-
- Clevers H, Nusse R. Wnt/β-Catenin Signaling and Disease. Cell. 2012;149:1192–1397. - PubMed
-
- Ng RC, Matsumaru D, Ho AS, Garcia-Barcelo MM, Yuan ZW, Smith D, Kodjabachian L, Tam PK, Yamada G, Lui VC. Dysregulation of Wnt inhibitory factor 1 (Wif1) expression resulted in aberrant Wnt-beta-catenin signaling and cell death of the cloaca endoderm, and anorectal malformations. Cell death and differentiation. 2014;21:978–989. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

