Approved Antiviral Drugs over the Past 50 Years
- PMID: 27281742
- PMCID: PMC4978613
- DOI: 10.1128/CMR.00102-15
Approved Antiviral Drugs over the Past 50 Years
Abstract
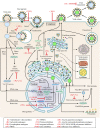
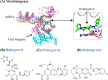
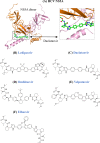
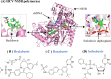
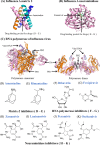
Since the first antiviral drug, idoxuridine, was approved in 1963, 90 antiviral drugs categorized into 13 functional groups have been formally approved for the treatment of the following 9 human infectious diseases: (i) HIV infections (protease inhibitors, integrase inhibitors, entry inhibitors, nucleoside reverse transcriptase inhibitors, nonnucleoside reverse transcriptase inhibitors, and acyclic nucleoside phosphonate analogues), (ii) hepatitis B virus (HBV) infections (lamivudine, interferons, nucleoside analogues, and acyclic nucleoside phosphonate analogues), (iii) hepatitis C virus (HCV) infections (ribavirin, interferons, NS3/4A protease inhibitors, NS5A inhibitors, and NS5B polymerase inhibitors), (iv) herpesvirus infections (5-substituted 2'-deoxyuridine analogues, entry inhibitors, nucleoside analogues, pyrophosphate analogues, and acyclic guanosine analogues), (v) influenza virus infections (ribavirin, matrix 2 protein inhibitors, RNA polymerase inhibitors, and neuraminidase inhibitors), (vi) human cytomegalovirus infections (acyclic guanosine analogues, acyclic nucleoside phosphonate analogues, pyrophosphate analogues, and oligonucleotides), (vii) varicella-zoster virus infections (acyclic guanosine analogues, nucleoside analogues, 5-substituted 2'-deoxyuridine analogues, and antibodies), (viii) respiratory syncytial virus infections (ribavirin and antibodies), and (ix) external anogenital warts caused by human papillomavirus infections (imiquimod, sinecatechins, and podofilox). Here, we present for the first time a comprehensive overview of antiviral drugs approved over the past 50 years, shedding light on the development of effective antiviral treatments against current and emerging infectious diseases worldwide.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
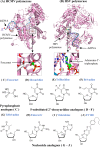
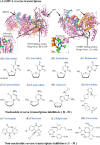
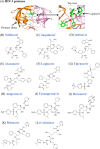
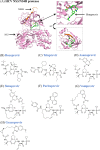




Similar articles
-
Antiviral drugs in current clinical use.J Clin Virol. 2004 Jun;30(2):115-33. doi: 10.1016/j.jcv.2004.02.009. J Clin Virol. 2004. PMID: 15125867 Review.
-
Another ten stories in antiviral drug discovery (part C): "Old" and "new" antivirals, strategies, and perspectives.Med Res Rev. 2009 Jul;29(4):611-45. doi: 10.1002/med.20153. Med Res Rev. 2009. PMID: 19260077 Review.
-
Drug Discovery of Nucleos(t)ide Antiviral Agents: Dedicated to Prof. Dr. Erik De Clercq on Occasion of His 80th Birthday.Molecules. 2021 Feb 9;26(4):923. doi: 10.3390/molecules26040923. Molecules. 2021. PMID: 33572409 Free PMC article. Review.
-
Emerging antiviral drugs.Expert Opin Emerg Drugs. 2008 Sep;13(3):393-416. doi: 10.1517/14728214.13.3.393. Expert Opin Emerg Drugs. 2008. PMID: 18764719 Review.
-
Antiviral drugs for viruses other than human immunodeficiency virus.Mayo Clin Proc. 2011 Oct;86(10):1009-26. doi: 10.4065/mcp.2011.0309. Mayo Clin Proc. 2011. PMID: 21964179 Free PMC article. Review.
Cited by
-
COVID-19 and SARS-CoV-2. Modeling the present, looking at the future.Phys Rep. 2020 Jul 10;869:1-51. doi: 10.1016/j.physrep.2020.07.005. Epub 2020 Jul 28. Phys Rep. 2020. PMID: 32834430 Free PMC article. Review.
-
A review: Mechanism of action of antiviral drugs.Int J Immunopathol Pharmacol. 2021 Jan-Dec;35:20587384211002621. doi: 10.1177/20587384211002621. Int J Immunopathol Pharmacol. 2021. PMID: 33726557 Free PMC article. Review.
-
Synthesis, characterization, molecular docking, and anticancer activities of new 1,3,4-oxadiazole-5-fluorocytosine hybrid derivatives.J Mol Struct. 2022 Sep 9:134135. doi: 10.1016/j.molstruc.2022.134135. Online ahead of print. J Mol Struct. 2022. PMID: 36101881 Free PMC article.
-
HIV-Induced Thymic Insufficiency and Aging-Related Immunosenescence on Immune Reconstitution in ART-Treated Patients.Vaccines (Basel). 2024 Jun 4;12(6):612. doi: 10.3390/vaccines12060612. Vaccines (Basel). 2024. PMID: 38932341 Free PMC article.
-
Cardiovascular adverse effects of antiviral therapies for COVID-19: Evidence and plausible mechanisms.Acta Pharmacol Sin. 2025 Mar;46(3):554-564. doi: 10.1038/s41401-024-01382-w. Epub 2024 Sep 9. Acta Pharmacol Sin. 2025. PMID: 39251859 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

