Virion Structure of Iflavirus Slow Bee Paralysis Virus at 2.6-Angstrom Resolution
- PMID: 27279610
- PMCID: PMC4984619
- DOI: 10.1128/JVI.00680-16
Virion Structure of Iflavirus Slow Bee Paralysis Virus at 2.6-Angstrom Resolution
Abstract
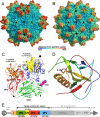
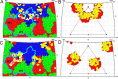
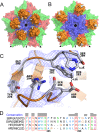
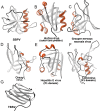
The western honeybee (Apis mellifera) is the most important commercial insect pollinator. However, bees are under pressure from habitat loss, environmental stress, and pathogens, including viruses that can cause lethal epidemics. Slow bee paralysis virus (SBPV) belongs to the Iflaviridae family of nonenveloped single-stranded RNA viruses. Here we present the structure of the SBPV virion determined from two crystal forms to resolutions of 3.4 Å and 2.6 Å. The overall structure of the virion resembles that of picornaviruses, with the three major capsid proteins VP1 to 3 organized into a pseudo-T3 icosahedral capsid. However, the SBPV capsid protein VP3 contains a C-terminal globular domain that has not been observed in other viruses from the order Picornavirales The protruding (P) domains form "crowns" on the virion surface around each 5-fold axis in one of the crystal forms. However, the P domains are shifted 36 Å toward the 3-fold axis in the other crystal form. Furthermore, the P domain contains the Ser-His-Asp triad within a surface patch of eight conserved residues that constitutes a putative catalytic or receptor-binding site. The movements of the domain might be required for efficient substrate cleavage or receptor binding during virus cell entry. In addition, capsid protein VP2 contains an RGD sequence that is exposed on the virion surface, indicating that integrins might be cellular receptors of SBPV.
Importance: Pollination by honeybees is needed to sustain agricultural productivity as well as the biodiversity of wild flora. However, honeybee populations in Europe and North America have been declining since the 1950s. Honeybee viruses from the Iflaviridae family are among the major causes of honeybee colony mortality. We determined the virion structure of an Iflavirus, slow bee paralysis virus (SBPV). SBPV exhibits unique structural features not observed in other picorna-like viruses. The SBPV capsid protein VP3 has a large C-terminal domain, five of which form highly prominent protruding "crowns" on the virion surface. However, the domains can change their positions depending on the conditions of the environment. The domain includes a putative catalytic or receptor binding site that might be important for SBPV cell entry.
Copyright © 2016 Kalynych et al.
Figures

Similar articles
-
Virion Structure of Israeli Acute Bee Paralysis Virus.J Virol. 2016 Aug 26;90(18):8150-9. doi: 10.1128/JVI.00854-16. Print 2016 Sep 15. J Virol. 2016. PMID: 27384649 Free PMC article.
-
Structure of deformed wing virus, a major honey bee pathogen.Proc Natl Acad Sci U S A. 2017 Mar 21;114(12):3210-3215. doi: 10.1073/pnas.1615695114. Epub 2017 Mar 7. Proc Natl Acad Sci U S A. 2017. PMID: 28270616 Free PMC article.
-
Virion Structure of Black Queen Cell Virus, a Common Honeybee Pathogen.J Virol. 2017 Feb 28;91(6):e02100-16. doi: 10.1128/JVI.02100-16. Print 2017 Mar 15. J Virol. 2017. PMID: 28077635 Free PMC article.
-
Virion structures and genome delivery of honeybee viruses.Curr Opin Virol. 2020 Dec;45:17-24. doi: 10.1016/j.coviro.2020.06.007. Epub 2020 Jul 14. Curr Opin Virol. 2020. PMID: 32679289 Review.
-
Functional aspects of the capsid structure of Mengo virus.J Struct Biol. 1990 Jul-Sep;104(1-3):52-62. doi: 10.1016/1047-8477(90)90057-j. J Struct Biol. 1990. PMID: 1965133 Review.
Cited by
-
The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.Commun Biol. 2019 Feb 20;2:72. doi: 10.1038/s42003-019-0311-z. eCollection 2019. Commun Biol. 2019. PMID: 30820467 Free PMC article.
-
Pathogens Spillover from Honey Bees to Other Arthropods.Pathogens. 2021 Aug 17;10(8):1044. doi: 10.3390/pathogens10081044. Pathogens. 2021. PMID: 34451508 Free PMC article. Review.
-
Capsid Structure of a Marine Algal Virus of the Order Picornavirales.J Virol. 2020 Apr 16;94(9):e01855-19. doi: 10.1128/JVI.01855-19. Print 2020 Apr 16. J Virol. 2020. PMID: 32024776 Free PMC article.
-
Capsid opening enables genome release of iflaviruses.Sci Adv. 2021 Jan 1;7(1):eabd7130. doi: 10.1126/sciadv.abd7130. Print 2021 Jan. Sci Adv. 2021. PMID: 33523856 Free PMC article.
-
A Novel Iflavirus Was Discovered in Green Rice Leafhopper Nephotettix cincticeps and Its Proliferation Was Inhibited by Infection of Rice Dwarf Virus.Front Microbiol. 2021 Jan 8;11:621141. doi: 10.3389/fmicb.2020.621141. eCollection 2020. Front Microbiol. 2021. PMID: 33488564 Free PMC article.
References
-
- Biesmeijer JC, Roberts SP, Reemer M, Ohlemuller R, Edwards M, Peeters T, Schaffers AP, Potts SG, Kleukers R, Thomas CD, Settele J, Kunin WE. 2006. Parallel declines in pollinators and insect-pollinated plants in Britain and the Netherlands. Science 313:351–354. doi:10.1126/science.1127863. - DOI - PubMed
-
- Vanengelsdorp D, Meixner MD. 2010. A historical review of managed honey bee populations in Europe and the United States and the factors that may affect them. J Invertebr Pathol 103(Suppl 1):S80–S95. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

