Network representation of protein interactions: Theory of graph description and analysis
- PMID: 27272236
- PMCID: PMC5338233
- DOI: 10.1002/pro.2963
Network representation of protein interactions: Theory of graph description and analysis
Abstract
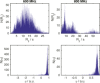
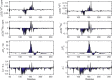
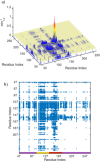
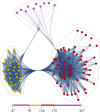
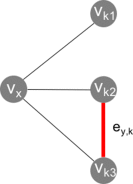
A methodological framework is presented for the graph theoretical interpretation of NMR data of protein interactions. The proposed analysis generalizes the idea of network representations of protein structures by expanding it to protein interactions. This approach is based on regularization of residue-resolved NMR relaxation times and chemical shift data and subsequent construction of an adjacency matrix that represents the underlying protein interaction as a graph or network. The network nodes represent protein residues. Two nodes are connected if two residues are functionally correlated during the protein interaction event. The analysis of the resulting network enables the quantification of the importance of each amino acid of a protein for its interactions. Furthermore, the determination of the pattern of correlations between residues yields insights into the functional architecture of an interaction. This is of special interest for intrinsically disordered proteins, since the structural (three-dimensional) architecture of these proteins and their complexes is difficult to determine. The power of the proposed methodology is demonstrated at the example of the interaction between the intrinsically disordered protein osteopontin and its natural ligand heparin.
Keywords: chemical shift; graph theory; network description; nuclear magnetic resonance; protein interactions; relaxation.
© 2016 The Protein Society.
Figures
Similar articles
-
NMR probing and visualization of correlated structural fluctuations in intrinsically disordered proteins.Phys Chem Chem Phys. 2017 Apr 19;19(16):10651-10656. doi: 10.1039/c7cp00430c. Phys Chem Chem Phys. 2017. PMID: 28397898
-
Network representation of protein interactions-Experimental results.Protein Sci. 2016 Sep;25(9):1628-36. doi: 10.1002/pro.2964. Epub 2016 Jun 16. Protein Sci. 2016. PMID: 27272395 Free PMC article.
-
Transient protein-protein interactions visualized by solution NMR.Biochim Biophys Acta. 2016 Jan;1864(1):115-22. doi: 10.1016/j.bbapap.2015.04.009. Epub 2015 Apr 18. Biochim Biophys Acta. 2016. PMID: 25896389 Review.
-
A J-modulated protonless NMR experiment characterizes the conformational ensemble of the intrinsically disordered protein WIP.J Biomol NMR. 2016 Dec;66(4):243-257. doi: 10.1007/s10858-016-0073-6. Epub 2016 Nov 14. J Biomol NMR. 2016. PMID: 27844185
-
The multifaceted roles of intrinsic disorder in protein complexes.FEBS Lett. 2015 Sep 14;589(19 Pt A):2498-506. doi: 10.1016/j.febslet.2015.06.004. Epub 2015 Jun 11. FEBS Lett. 2015. PMID: 26073257 Review.
Cited by
-
Molecular Dynamics Simulations of Kir2.2 Interactions with an Ensemble of Cholesterol Molecules.Biophys J. 2018 Oct 2;115(7):1264-1280. doi: 10.1016/j.bpj.2018.07.041. Epub 2018 Aug 23. Biophys J. 2018. PMID: 30205899 Free PMC article.
-
Effects of pH on an IDP conformational ensemble explored by molecular dynamics simulation.Biophys Chem. 2021 Apr;271:106552. doi: 10.1016/j.bpc.2021.106552. Epub 2021 Jan 26. Biophys Chem. 2021. PMID: 33581430 Free PMC article.
-
Protein conformational switch discerned via network centrality properties.Comput Struct Biotechnol J. 2021 Jun 5;19:3599-3608. doi: 10.1016/j.csbj.2021.06.004. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34257839 Free PMC article.
-
Pervasive epistasis exposes intramolecular networks in adaptive enzyme evolution.Nat Commun. 2023 Dec 21;14(1):8508. doi: 10.1038/s41467-023-44333-5. Nat Commun. 2023. PMID: 38129396 Free PMC article.
-
Chokepoints in Mechanical Coupling Associated with Allosteric Proteins: The Pyruvate Kinase Example.Biophys J. 2019 May 7;116(9):1598-1608. doi: 10.1016/j.bpj.2019.03.026. Epub 2019 Apr 2. Biophys J. 2019. PMID: 31010662 Free PMC article.
References
-
- Jensen MR, Ruigrok RWH, Blackledge M (2013) Describing intrinsically disordered proteins at atomic resolution by NMR. Curr Opin Struct Biol 23:426–435. - PubMed
-
- Korzhnev DM, Kay LE (2008) Probing invisible, low‐populated states of protein molecules by relaxation dispersion NMR spectroscopy: an application to protein folding. Acc Chem Res 41:442–451. - PubMed
-
- Rule GS, Hitchens TK ( 2006) Fundamentals of protein NMR spectroscopy. Dordrecht: Springer.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

