Opportunistic intruders: how viruses orchestrate ER functions to infect cells
- PMID: 27265768
- PMCID: PMC5272919
- DOI: 10.1038/nrmicro.2016.60
Opportunistic intruders: how viruses orchestrate ER functions to infect cells
Abstract
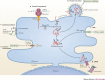
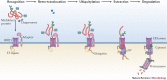
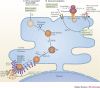
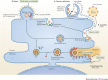
Viruses subvert the functions of their host cells to replicate and form new viral progeny. The endoplasmic reticulum (ER) has been identified as a central organelle that governs the intracellular interplay between viruses and hosts. In this Review, we analyse how viruses from vastly different families converge on this unique intracellular organelle during infection, co-opting some of the endogenous functions of the ER to promote distinct steps of the viral life cycle from entry and replication to assembly and egress. The ER can act as the common denominator during infection for diverse virus families, thereby providing a shared principle that underlies the apparent complexity of relationships between viruses and host cells. As a plethora of information illuminating the molecular and cellular basis of virus-ER interactions has become available, these insights may lead to the development of crucial therapeutic agents.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
Similar articles
-
ER functions are exploited by viruses to support distinct stages of their life cycle.Biochem Soc Trans. 2020 Oct 30;48(5):2173-2184. doi: 10.1042/BST20200395. Biochem Soc Trans. 2020. PMID: 33119046 Free PMC article. Review.
-
How viruses use the endoplasmic reticulum for entry, replication, and assembly.Cold Spring Harb Perspect Biol. 2013 Jan 1;5(1):a013250. doi: 10.1101/cshperspect.a013250. Cold Spring Harb Perspect Biol. 2013. PMID: 23284050 Free PMC article. Review.
-
Endoplasmic Reticulum: The Favorite Intracellular Niche for Viral Replication and Assembly.Viruses. 2016 Jun 7;8(6):160. doi: 10.3390/v8060160. Viruses. 2016. PMID: 27338443 Free PMC article. Review.
-
Endoplasmic reticulum in viral infection.Int Rev Cell Mol Biol. 2020;350:265-284. doi: 10.1016/bs.ircmb.2019.10.005. Epub 2019 Nov 8. Int Rev Cell Mol Biol. 2020. PMID: 32138901 Review.
-
How Viruses Use the VCP/p97 ATPase Molecular Machine.Viruses. 2021 Sep 21;13(9):1881. doi: 10.3390/v13091881. Viruses. 2021. PMID: 34578461 Free PMC article. Review.
Cited by
-
Small GTPase-A Key Role in Host Cell for Coronavirus Infection and a Potential Target for Coronavirus Vaccine Adjuvant Discovery.Viruses. 2022 Sep 14;14(9):2044. doi: 10.3390/v14092044. Viruses. 2022. PMID: 36146850 Free PMC article. Review.
-
Eating the unknown: Xenophagy and ER-phagy are cytoprotective defenses against pathogens.Exp Cell Res. 2020 Nov 1;396(1):112276. doi: 10.1016/j.yexcr.2020.112276. Epub 2020 Sep 9. Exp Cell Res. 2020. PMID: 32918896 Free PMC article. Review.
-
The astrovirus N-terminal nonstructural protein anchors replication complexes to the perinuclear ER membranes.PLoS Pathog. 2024 Jul 15;20(7):e1011959. doi: 10.1371/journal.ppat.1011959. eCollection 2024 Jul. PLoS Pathog. 2024. PMID: 39008516 Free PMC article.
-
Dengue virus exploits the host tRNA epitranscriptome to promote viral replication.bioRxiv [Preprint]. 2023 Nov 6:2023.11.05.565734. doi: 10.1101/2023.11.05.565734. bioRxiv. 2023. PMID: 37986976 Free PMC article. Preprint.
-
Atlastin Endoplasmic Reticulum-Shaping Proteins Facilitate Zika Virus Replication.J Virol. 2019 Nov 13;93(23):e01047-19. doi: 10.1128/JVI.01047-19. Print 2019 Dec 1. J Virol. 2019. PMID: 31534046 Free PMC article.
References
-
- Shibata Y, Voeltz GK, Rapoport TA. Rough sheets and smooth tubules. Cell. 2006;126:435–439. - PubMed
-
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. - PubMed
-
- Westrate LM, Lee JE, Prinz WA, Voeltz GK. Form follows function: the importance of endoplasmic reticulum shape. Annu. Rev. Biochem. 2015;84:791–811. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

