Immunological and clinical significance of HLA class I antigen processing machinery component defects in malignant cells
- PMID: 27264839
- PMCID: PMC5264518
- DOI: 10.1016/j.oraloncology.2016.05.008
Immunological and clinical significance of HLA class I antigen processing machinery component defects in malignant cells
Abstract
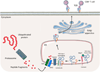
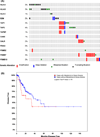
Experimental as well as clinical studies demonstrate that the immune system plays a major role in controlling generation and progression of tumors. The cancer immunoediting theory supports the notion that tumor cell immunogenicity is dynamically shaped by the immune system, as it eliminates immunogenic tumor cells in the early stage of the disease and then edits their antigenicity. The end result is the generation of a tumor cell population able to escape from immune recognition and elimination by tumor infiltrating lymphocytes. Two major mechanisms, which affect the target cells and the effector phase of the immune response, play a crucial role in the editing process. One is represented by the downregulation of tumor antigen (TA) processing and presentation because of abnormalities in the HLA class I antigen processing machinery (APM). The other one is represented by the anergy of effector immune infiltrates in the tumor microenvironment caused by aberrant inhibitory signals triggered by immune checkpoint receptor (ICR) ligands, such as programmed death ligand-1 (PD-L1). In this review, we will focus on tumor immune escape mechanisms caused by defects in HLA class I APM component expression and/or function in different types of cancer, with emphasis on head and neck cancer (HNC). We will also discuss the immunological implications and clinical relevance of these HLA class I APM abnormalities. Finally, we will describe strategies to counteract defective TA presentation with the expectation that they will enhance tumor recognition and elimination by tumor infiltrating effector T cells.
Keywords: APM; CTL; HLA class I; Immunoescape.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Conflict of interest statement
statement The authors disclose no potential conflicts of interest.
Figures
Similar articles
-
HLA class I antigen processing machinery (APM) component expression and PD-1:PD-L1 pathway activation in HIV-infected head and neck cancers.Oral Oncol. 2018 Feb;77:92-97. doi: 10.1016/j.oraloncology.2017.12.014. Epub 2017 Dec 28. Oral Oncol. 2018. PMID: 29362132 Free PMC article.
-
Immune escape associated with functional defects in antigen-processing machinery in head and neck cancer.Clin Cancer Res. 2006 Jul 1;12(13):3890-5. doi: 10.1158/1078-0432.CCR-05-2750. Clin Cancer Res. 2006. PMID: 16818683 Review.
-
STAT1-Induced HLA Class I Upregulation Enhances Immunogenicity and Clinical Response to Anti-EGFR mAb Cetuximab Therapy in HNC Patients.Cancer Immunol Res. 2015 Aug;3(8):936-45. doi: 10.1158/2326-6066.CIR-15-0053. Epub 2015 May 13. Cancer Immunol Res. 2015. PMID: 25972070 Free PMC article.
-
Expression and functional analysis of human leukocyte antigen class I antigen-processing machinery in medulloblastoma.Cancer Res. 2007 Jun 1;67(11):5471-8. doi: 10.1158/0008-5472.CAN-06-4735. Cancer Res. 2007. PMID: 17545629
-
Human leukocyte antigen (HLA) class I defects in head and neck cancer: molecular mechanisms and clinical significance.Immunol Res. 2005;33(2):113-33. doi: 10.1385/IR:33:2:113. Immunol Res. 2005. PMID: 16234579 Review.
Cited by
-
Immune-Related Mutational Landscape and Gene Signatures: Prognostic Value and Therapeutic Impact for Head and Neck Cancer.Cancers (Basel). 2021 Mar 8;13(5):1162. doi: 10.3390/cancers13051162. Cancers (Basel). 2021. PMID: 33800421 Free PMC article. Review.
-
Heterogeneity of HLA-G Expression in Cancers: Facing the Challenges.Front Immunol. 2018 Sep 27;9:2164. doi: 10.3389/fimmu.2018.02164. eCollection 2018. Front Immunol. 2018. PMID: 30319626 Free PMC article. Review.
-
The Potential of Soluble Human Leukocyte Antigen Molecules for Early Cancer Detection and Therapeutic Vaccine Design.Vaccines (Basel). 2020 Dec 18;8(4):775. doi: 10.3390/vaccines8040775. Vaccines (Basel). 2020. PMID: 33353014 Free PMC article. Review.
-
Mechanisms of Resistance and Therapeutic Perspectives in Immunotherapy for Advanced Head and Neck Cancers.Cancers (Basel). 2024 Feb 7;16(4):703. doi: 10.3390/cancers16040703. Cancers (Basel). 2024. PMID: 38398094 Free PMC article. Review.
-
DNA Vaccines-How Far From Clinical Use?Int J Mol Sci. 2018 Nov 15;19(11):3605. doi: 10.3390/ijms19113605. Int J Mol Sci. 2018. PMID: 30445702 Free PMC article. Review.
References
-
- Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–998. - PubMed
-
- Ostrand-Rosenberg S. Animal models of tumor immunity, immunotherapy and cancer vaccines. Curr Opin Immunol. 2004;16:143–150. - PubMed
-
- Marsman M, Jordens I, Griekspoor A, Neefjes J. Chaperoning antigen presentation by MHC class II molecules and their role in oncogenesis. Adv Cancer Res. 2005;93:129–158. - PubMed
-
- Moretta L, Bottino C, Pende D, Vitale M, Mingari MC, Moretta A. Human natural killer cells: molecular mechanisms controlling NK cell activation and tumor cell lysis. Immunol Lett. 2005;100:7–13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

