Compartmentation of metabolites in regulating epigenome of cancer
- PMID: 27258652
- PMCID: PMC5072407
- DOI: 10.2119/molmed.2016.00051
Compartmentation of metabolites in regulating epigenome of cancer
Abstract
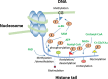
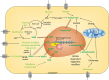
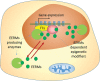
Covalent modification of DNA and histones are important epigenetic events and the genome wide reshaping of epigenetic markers is common in cancer. The epigenetic markers are produced by enzymatic reactions and some of these reactions require the presence of metabolites as cofactors (termed Epigenetic Enzyme Required Metabolites, EERMs). Recent studies found that the abundance of these EERMs correlates with epigenetic enzyme activities. Also, the subcellular compartmentation, especially the nuclear localization of these EERMs may play a role in regulating the activities of epigenetic enzymes. Moreover, gene specific recruitment of enzymes which produce the EERMs in the proximity of the epigenetic modification events accompanying the gene expression regulation, were proposed. Therefore, it is of importance to summarize these findings of the EERMs in regulating the epigenetic modifications at both DNA and histone levels, and to understand how EERMs contribute to cancer development by addressing their global versus local distribution.
Keywords: DNA methylation; EERM; Epigenetics; cancer cell metabolism; gene regulation; histone modification; metabolites.
Conflict of interest statement
The authors declare that they have no competing interests as defined by
Figures
Similar articles
-
Cancer chemoprevention and nutriepigenetics: state of the art and future challenges.Top Curr Chem. 2013;329:73-132. doi: 10.1007/128_2012_360. Top Curr Chem. 2013. PMID: 22955508 Review.
-
Interplay between different epigenetic modifications and mechanisms.Adv Genet. 2010;70:101-41. doi: 10.1016/B978-0-12-380866-0.60005-8. Adv Genet. 2010. PMID: 20920747 Review.
-
[Epigenome and cancer: new possibilities of cancer prevention and therapy?].Postepy Biochem. 2005;51(3):244-50. Postepy Biochem. 2005. PMID: 16381168 Review. Polish.
-
DNA methylation, histone acetylation and methylation of epigenetic modifications as a therapeutic approach for cancers.Cancer Lett. 2016 Apr 10;373(2):185-92. doi: 10.1016/j.canlet.2016.01.036. Epub 2016 Jan 22. Cancer Lett. 2016. PMID: 26808576 Review.
-
Epigenetic drift towards histone modifications regulates CAV1 gene expression in colon cancer.Gene. 2016 Apr 25;581(1):75-84. doi: 10.1016/j.gene.2016.01.029. Epub 2016 Jan 19. Gene. 2016. PMID: 26794448
Cited by
-
Metabolic Dysregulations and Epigenetics: A Bidirectional Interplay that Drives Tumor Progression.Cells. 2019 Jul 30;8(8):798. doi: 10.3390/cells8080798. Cells. 2019. PMID: 31366176 Free PMC article. Review.
-
Methylome Variation Predicts Exemestane Resistance in Advanced ER+ Breast Cancer.Technol Cancer Res Treat. 2020 Jan-Dec;19:1533033819896331. doi: 10.1177/1533033819896331. Technol Cancer Res Treat. 2020. PMID: 32129154 Free PMC article.
-
Molecular link between glucose and glutamine consumption in cancer cells mediated by CtBP and SIRT4.Oncogenesis. 2018 Mar 13;7(3):26. doi: 10.1038/s41389-018-0036-8. Oncogenesis. 2018. PMID: 29540733 Free PMC article.
-
VDAC1 is a molecular target in glioblastoma, with its depletion leading to reprogrammed metabolism and reversed oncogenic properties.Neuro Oncol. 2017 Jul 1;19(7):951-964. doi: 10.1093/neuonc/now297. Neuro Oncol. 2017. PMID: 28339833 Free PMC article.
-
Mitochondrial VDAC1-based peptides: Attacking oncogenic properties in glioblastoma.Oncotarget. 2017 May 9;8(19):31329-31346. doi: 10.18632/oncotarget.15455. Oncotarget. 2017. PMID: 28412744 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

