C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector
- PMID: 27256883
- PMCID: PMC5127784
- DOI: 10.1126/science.aaf5573
C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector
Abstract
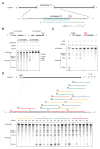
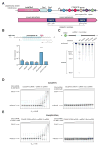
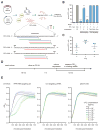
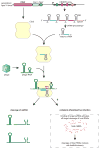
The clustered regularly interspaced short palindromic repeat (CRISPR)-CRISPR-associated genes (Cas) adaptive immune system defends microbes against foreign genetic elements via DNA or RNA-DNA interference. We characterize the class 2 type VI CRISPR-Cas effector C2c2 and demonstrate its RNA-guided ribonuclease function. C2c2 from the bacterium Leptotrichia shahii provides interference against RNA phage. In vitro biochemical analysis shows that C2c2 is guided by a single CRISPR RNA and can be programmed to cleave single-stranded RNA targets carrying complementary protospacers. In bacteria, C2c2 can be programmed to knock down specific mRNAs. Cleavage is mediated by catalytic residues in the two conserved Higher Eukaryotes and Prokaryotes Nucleotide-binding (HEPN) domains, mutations of which generate catalytically inactive RNA-binding proteins. These results broaden our understanding of CRISPR-Cas systems and suggest that C2c2 can be used to develop new RNA-targeting tools.
Copyright © 2016, American Association for the Advancement of Science.
Figures



Similar articles
-
Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection.Nature. 2016 Oct 13;538(7624):270-273. doi: 10.1038/nature19802. Epub 2016 Sep 26. Nature. 2016. PMID: 27669025 Free PMC article.
-
Molecular Mechanisms of RNA Targeting by Cas13-containing Type VI CRISPR-Cas Systems.J Mol Biol. 2019 Jan 4;431(1):66-87. doi: 10.1016/j.jmb.2018.06.029. Epub 2018 Jun 22. J Mol Biol. 2019. PMID: 29940185 Review.
-
Structural basis for self-cleavage prevention by tag:anti-tag pairing complementarity in type VI Cas13 CRISPR systems.Mol Cell. 2021 Mar 4;81(5):1100-1115.e5. doi: 10.1016/j.molcel.2020.12.033. Epub 2021 Jan 19. Mol Cell. 2021. PMID: 33472057 Free PMC article.
-
The Revolution Continues: Newly Discovered Systems Expand the CRISPR-Cas Toolkit.Mol Cell. 2017 Oct 5;68(1):15-25. doi: 10.1016/j.molcel.2017.09.007. Mol Cell. 2017. PMID: 28985502 Free PMC article. Review.
-
Approaches to study CRISPR RNA biogenesis and the key players involved.Methods. 2020 Feb 1;172:12-26. doi: 10.1016/j.ymeth.2019.07.015. Epub 2019 Jul 17. Methods. 2020. PMID: 31325492 Review.
Cited by
-
A resurrected ancestor of Cas12a expands target access and substrate recognition for nucleic acid editing and detection.Nat Biotechnol. 2024 Oct 31. doi: 10.1038/s41587-024-02461-3. Online ahead of print. Nat Biotechnol. 2024. PMID: 39482449
-
Using Drosophila to uncover molecular and physiological functions of circRNAs.Methods. 2021 Dec;196:74-84. doi: 10.1016/j.ymeth.2021.04.016. Epub 2021 Apr 24. Methods. 2021. PMID: 33901645 Free PMC article. Review.
-
How to measure and evaluate binding affinities.Elife. 2020 Aug 6;9:e57264. doi: 10.7554/eLife.57264. Elife. 2020. PMID: 32758356 Free PMC article.
-
MeCas12a, a Highly Sensitive and Specific System for COVID-19 Detection.Adv Sci (Weinh). 2020 Sep 23;7(20):2001300. doi: 10.1002/advs.202001300. eCollection 2020 Oct. Adv Sci (Weinh). 2020. PMID: 33042732 Free PMC article.
-
Compact RNA editors with natural miniature Cas13j nucleases.Nat Chem Biol. 2024 Sep 19. doi: 10.1038/s41589-024-01729-8. Online ahead of print. Nat Chem Biol. 2024. PMID: 39300230
References
-
- Wright AV, Nunez JK, Doudna JA. Biology and Applications of CRISPR Systems: Harnessing Nature's Toolbox for Genome Engineering. Cell. 2016;164:29–44. - PubMed
-
- Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61. - PubMed
-
- van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci. 2009;34:401–407. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

