Genomic survey sequencing for development and validation of single-locus SSR markers in peanut (Arachis hypogaea L.)
- PMID: 27251557
- PMCID: PMC4888616
- DOI: 10.1186/s12864-016-2743-x
Genomic survey sequencing for development and validation of single-locus SSR markers in peanut (Arachis hypogaea L.)
Abstract
Background: Single-locus markers have many advantages compared with multi-locus markers in genetic and breeding studies because their alleles can be assigned to particular genomic loci in diversity analyses. However, there is little research on single-locus SSR markers in peanut. Through the de novo assembly of DNA sequencing reads of A. hypogaea, we developed single-locus SSR markers in a genomic survey for better application in genetic and breeding studies of peanut.
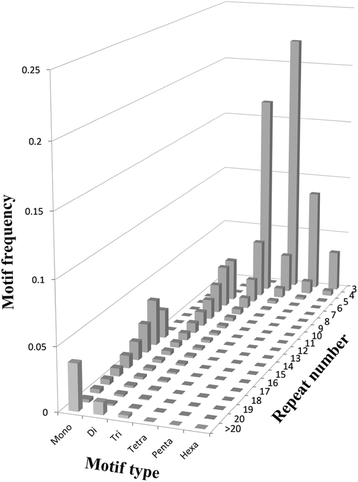
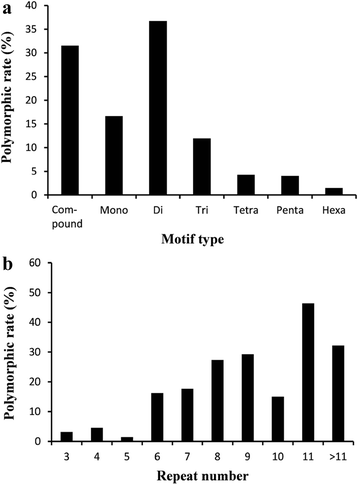
Results: In this study, DNA libraries with four different insert sizes were used for sequencing with 150 bp paired-end reads. Approximately 237 gigabases of clean data containing 1,675,631,984 reads were obtained after filtering. These reads were assembled into 2,102,446 contigs with an N50 length of 1,782 bp, and the contigs were further assembled into 1,176,527 scaffolds with an N50 of 3,920 bp. The total length of the assembled scaffold sequences was 2.0 Gbp, and 134,652 single-locus SSRs were identified from 375,180 SSRs. Among these developed single-locus SSRs, trinucleotide motifs were the most abundant, followed by tetra-, di-, mono-, penta- and hexanucleotide motifs. The most common motif repeats for the various types of single-locus SSRs have a tendency to be A/T rich. A total of 1,790 developed in silico single-locus SSR markers were chosen and used in PCR experiments to confirm amplification patterns. Of them, 1,637 markers that produced single amplicons in twelve inbred lines were considered putative single-locus markers, and 290 (17.7 %) showed polymorphisms. A further F2 population study showed that the segregation ratios of the 97 developed SSR markers, which showed polymorphisms between the parents, were consistent with the Mendelian inheritance law for single loci (1:2:1). Finally, 89 markers were assigned to an A. hypogaea linkage map. A subset of 100 single-locus SSR markers was shown to be highly stable and universal in a collection of 96 peanut accessions. A neighbor-joining tree of this natural population showed that genotypes have obviously correlation with botanical varieties.
Conclusions: We have shown that the detection of single-locus SSR markers from a de novo genomic assembly of a combination of different-insert-size libraries is highly efficient. This is the first report of the development of genome-wide single-locus markers for A. hypogaea, and the markers developed in this study will be useful for gene tagging, sequence scaffold assignment, linkage map construction, diversity analysis, variety identification and association mapping in peanut.
Keywords: Combined libraries; Peanut (A hypogaea L.); Single-locus SSR; de novo assembly.
Figures

Similar articles
-
Molecular marker development from transcript sequences and germplasm evaluation for cultivated peanut (Arachis hypogaea L.).Mol Genet Genomics. 2016 Feb;291(1):363-81. doi: 10.1007/s00438-015-1115-6. Epub 2015 Sep 11. Mol Genet Genomics. 2016. PMID: 26362763
-
Genetic diversity of peanut (Arachis hypogaea L.) and its wild relatives based on the analysis of hypervariable regions of the genome.BMC Plant Biol. 2004 Jul 14;4:11. doi: 10.1186/1471-2229-4-11. BMC Plant Biol. 2004. PMID: 15253775 Free PMC article.
-
Genome-wide identification of microsatellite markers from cultivated peanut (Arachis hypogaea L.).BMC Genomics. 2019 Nov 1;20(1):799. doi: 10.1186/s12864-019-6148-5. BMC Genomics. 2019. PMID: 31675924 Free PMC article.
-
Advances in genetics and molecular breeding of three legume crops of semi-arid tropics using next-generation sequencing and high-throughput genotyping technologies.J Biosci. 2012 Nov;37(5):811-20. doi: 10.1007/s12038-012-9228-0. J Biosci. 2012. PMID: 23107917 Review.
-
Advances in the evolution research and genetic breeding of peanut.Gene. 2024 Jul 20;916:148425. doi: 10.1016/j.gene.2024.148425. Epub 2024 Apr 3. Gene. 2024. PMID: 38575102 Review.
Cited by
-
High-throughput SSR marker development and its application in a centipedegrass (Eremochloa ophiuroides (Munro) Hack.) genetic diversity analysis.PLoS One. 2018 Aug 22;13(8):e0202605. doi: 10.1371/journal.pone.0202605. eCollection 2018. PLoS One. 2018. PMID: 30133524 Free PMC article.
-
Co-localization of major quantitative trait loci for pod size and weight to a 3.7 cM interval on chromosome A05 in cultivated peanut (Arachis hypogaea L.).BMC Genomics. 2017 Jan 9;18(1):58. doi: 10.1186/s12864-016-3456-x. BMC Genomics. 2017. PMID: 28068921 Free PMC article.
-
Chromosomes A07 and A05 associated with stable and major QTLs for pod weight and size in cultivated peanut (Arachis hypogaea L.).Theor Appl Genet. 2018 Feb;131(2):267-282. doi: 10.1007/s00122-017-3000-7. Epub 2017 Oct 20. Theor Appl Genet. 2018. PMID: 29058050
-
Genetic Variation and Association Mapping of Seed-Related Traits in Cultivated Peanut (Arachis hypogaea L.) Using Single-Locus Simple Sequence Repeat Markers.Front Plant Sci. 2017 Dec 11;8:2105. doi: 10.3389/fpls.2017.02105. eCollection 2017. Front Plant Sci. 2017. PMID: 29321787 Free PMC article.
-
Genetic mapping of yield traits using RIL population derived from Fuchuan Dahuasheng and ICG6375 of peanut (Arachis hypogaea L.).Mol Breed. 2017;37(2):17. doi: 10.1007/s11032-016-0587-3. Epub 2017 Jan 30. Mol Breed. 2017. PMID: 28216998 Free PMC article.
References
-
- Kochert G, Stalker HT, Gimenes M, Galgaro L, Lopes CR, Moore K. RFLP and cytogenetic evidence on the origin and evolution of allotetraploid domesticated peanut, Arachis hypogaea (Leguminosae) Am J Bot. 1996;83:1282–1291. doi: 10.2307/2446112. - DOI
-
- Halward TM, Stalker HT, Larue EA, Kochert G. Genetic variation detectable with molecular markers among unadapted germplasm resources of cultivated peanut and related wild species. Genome. 1991;34:1013–1020. doi: 10.1139/g91-156. - DOI
-
- Burow MD, Simpson CE, Paterson AH, Starr JL. Identification of peanut (Arachishypogaea L.) RAPD markers diagnostic of root-knot nematode (Meloidogynearenaria (Neal) Chitwood) resistance. Mol Breeding. 1996;2:369–319. doi: 10.1007/BF00437915. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

