Sphingomyelin is sorted at the trans Golgi network into a distinct class of secretory vesicle
- PMID: 27247384
- PMCID: PMC4914164
- DOI: 10.1073/pnas.1602875113
Sphingomyelin is sorted at the trans Golgi network into a distinct class of secretory vesicle
Abstract
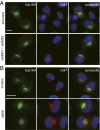
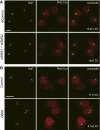
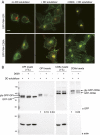
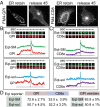
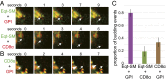
One of the principal functions of the trans Golgi network (TGN) is the sorting of proteins into distinct vesicular transport carriers that mediate secretion and interorganelle trafficking. Are lipids also sorted into distinct TGN-derived carriers? The Golgi is the principal site of the synthesis of sphingomyelin (SM), an abundant sphingolipid that is transported. To address the specificity of SM transport to the plasma membrane, we engineered a natural SM-binding pore-forming toxin, equinatoxin II (Eqt), into a nontoxic reporter termed Eqt-SM and used it to monitor intracellular trafficking of SM. Using quantitative live cell imaging, we found that Eqt-SM is enriched in a subset of TGN-derived secretory vesicles that are also enriched in a glycophosphatidylinositol-anchored protein. In contrast, an integral membrane secretory protein (CD8α) is not enriched in these carriers. Our results demonstrate the sorting of native SM at the TGN and its transport to the plasma membrane by specific carriers.
Keywords: Golgi apparatus; equinatoxin; secretion; sphingomyelin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Activity of the SPCA1 Calcium Pump Couples Sphingomyelin Synthesis to Sorting of Secretory Proteins in the Trans-Golgi Network.Dev Cell. 2018 Nov 19;47(4):464-478.e8. doi: 10.1016/j.devcel.2018.10.012. Epub 2018 Nov 1. Dev Cell. 2018. PMID: 30393074 Free PMC article.
-
Segregation of sphingolipids and sterols during formation of secretory vesicles at the trans-Golgi network.J Cell Biol. 2009 May 18;185(4):601-12. doi: 10.1083/jcb.200901145. Epub 2009 May 11. J Cell Biol. 2009. PMID: 19433450 Free PMC article.
-
Syndecan-1 Mediates Sorting of Soluble Lipoprotein Lipase with Sphingomyelin-Rich Membrane in the Golgi Apparatus.Dev Cell. 2019 Nov 4;51(3):387-398.e4. doi: 10.1016/j.devcel.2019.08.014. Epub 2019 Sep 19. Dev Cell. 2019. PMID: 31543446 Free PMC article.
-
Secretory cargo sorting at the trans-Golgi network.Trends Cell Biol. 2014 Oct;24(10):584-93. doi: 10.1016/j.tcb.2014.04.007. Epub 2014 May 16. Trends Cell Biol. 2014. PMID: 24841758 Review.
-
Phosphoinositides and membrane traffic at the trans-Golgi network.Biochem Soc Symp. 2005;(72):31-8. doi: 10.1042/bss0720031. Biochem Soc Symp. 2005. PMID: 15649127 Review.
Cited by
-
Chlamydia-containing spheres are a novel and predominant form of egress by the pathogen Chlamydia psittaci.mBio. 2024 Aug 14;15(8):e0128824. doi: 10.1128/mbio.01288-24. Epub 2024 Jul 23. mBio. 2024. PMID: 39041785 Free PMC article.
-
Ceramide Transfer Protein (CERT): An Overlooked Molecular Player in Cancer.Int J Mol Sci. 2021 Dec 7;22(24):13184. doi: 10.3390/ijms222413184. Int J Mol Sci. 2021. PMID: 34947980 Free PMC article. Review.
-
Direct trafficking pathways from the Golgi apparatus to the plasma membrane.Semin Cell Dev Biol. 2020 Nov;107:112-125. doi: 10.1016/j.semcdb.2020.04.001. Epub 2020 Apr 13. Semin Cell Dev Biol. 2020. PMID: 32317144 Free PMC article. Review.
-
Lipid Rafts: Controversies Resolved, Mysteries Remain.Trends Cell Biol. 2020 May;30(5):341-353. doi: 10.1016/j.tcb.2020.01.009. Epub 2020 Feb 20. Trends Cell Biol. 2020. PMID: 32302547 Free PMC article. Review.
-
INPP5E controls ciliary localization of phospholipids and the odor response in olfactory sensory neurons.J Cell Sci. 2022 Mar 1;135(5):jcs258364. doi: 10.1242/jcs.258364. Epub 2021 May 7. J Cell Sci. 2022. PMID: 33771931 Free PMC article.
References
-
- De Matteis MA, Luini A. Exiting the Golgi complex. Nat Rev Mol Cell Biol. 2008;9(4):273–284. - PubMed
-
- Litvak V, Dahan N, Ramachandran S, Sabanay H, Lev S. Maintenance of the diacylglycerol level in the Golgi apparatus by the Nir2 protein is critical for Golgi secretory function. Nat Cell Biol. 2005;7(3):225–234. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

