Stability of Reference Genes for Messenger RNA Quantification by Real-Time PCR in Mouse Dextran Sodium Sulfate Experimental Colitis
- PMID: 27244258
- PMCID: PMC4886971
- DOI: 10.1371/journal.pone.0156289
Stability of Reference Genes for Messenger RNA Quantification by Real-Time PCR in Mouse Dextran Sodium Sulfate Experimental Colitis
Abstract
Background: Many animal models have been developed to characterize the complexity of colonic inflammation. In dextran sodium sulfate (DSS) experimental colitis in mice the choice of reference genes is critical for accurate quantification of target genes using quantitative real time PCR (RT-qPCR). No studies have addressed the performance of reference genes in mice DSS-experimental colitis. This study aimed to determine the stability of reference genes expression (RGE) in DSS-experimental murine colitis.
Methods: Colitis was induced in male C57BL/6 mice using DSS5% for 5 days, control group received water. RNA was extracted from inflamed and non-inflamed colon. Using RT-qPCR, comparative analysis of 13 RGE was performed according to predefined criteria and relative colonic TNF-α and IL-1β gene expression was determined by calculating the difference in the threshold cycle.
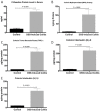
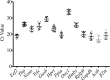
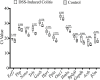
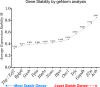
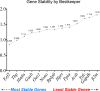
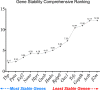
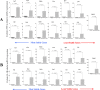
Results: Colitis significantly altered the stability of mucosal RGE. Commonly used glyceraldehyde-3-phosphate dehydrogenase (Gapdh), β-actin (Actb), or β2-microglobulin (β2m) showed the highest variability within the inflamed and control groups. Conversely, TATA-box-binding protein (Tbp) and eukaryotic translation elongation factor 2 (Eef2) were not affected by inflammation and were the most stable genes. Normalization of colonic TNF-α and IL-1β mRNA levels was dependent on the reference gene used. Depending on the genes used to normalize the data, statistical significance varied from significant when TBP / Eef2 were used to non-significant when Gapdh, Actb or β2m were used.
Conclusions: This study highlights the appropriate choice of RGE to ensure adequate normalization of RT-qPCR data when using this model. Suboptimal RGE may explain controversial results from published studies. We recommend using Tbp and Eef2 instead of Gapdh, Actb or β2m as reference genes.
Conflict of interest statement
Figures



Similar articles
-
Appropriateness of reference genes for normalizing messenger RNA in mouse 2,4-dinitrobenzene sulfonic acid (DNBS)-induced colitis using quantitative real time PCR.Sci Rep. 2017 Feb 10;7:42427. doi: 10.1038/srep42427. Sci Rep. 2017. PMID: 28186172 Free PMC article.
-
RNA Purity, Real-Time PCR Sensitivity, and Colon Segment Influence mRNA Relative Expression in Murine Dextran Sodium Sulfate Experimental Colitis.J Biomol Tech. 2018 Sep;29(3):61-70. doi: 10.7171/jbt.18-2903-001. Epub 2018 Jul 13. J Biomol Tech. 2018. PMID: 30034295 Free PMC article.
-
Orally administered glucans from the edible mushroom Pleurotus pulmonarius reduce acute inflammation in dextran sulfate sodium-induced experimental colitis.Br J Nutr. 2010 Feb;103(3):393-402. doi: 10.1017/S0007114509991760. Epub 2009 Sep 22. Br J Nutr. 2010. PMID: 19772681
-
Jianpi Qingchang decoction alleviates ulcerative colitis by inhibiting nuclear factor-κB activation.World J Gastroenterol. 2017 Feb 21;23(7):1180-1188. doi: 10.3748/wjg.v23.i7.1180. World J Gastroenterol. 2017. PMID: 28275298 Free PMC article.
-
Comparative study of candidate housekeeping genes for quantification of target gene messenger RNA expression by real-time PCR in patients with inflammatory bowel disease.Inflamm Bowel Dis. 2013 Dec;19(13):2840-7. doi: 10.1097/01.MIB.0000435440.22484.e8. Inflamm Bowel Dis. 2013. PMID: 24141710
Cited by
-
Denosumab Regulates Gut Microbiota Composition and Cytokines in Dinitrobenzene Sulfonic Acid (DNBS)-Experimental Colitis.Front Microbiol. 2020 Jun 25;11:1405. doi: 10.3389/fmicb.2020.01405. eCollection 2020. Front Microbiol. 2020. PMID: 32670246 Free PMC article.
-
Isolation and Characterization of Potentially Probiotic Bacterial Strains from Mice: Proof of Concept for Personalized Probiotics.Nutrients. 2018 Nov 5;10(11):1684. doi: 10.3390/nu10111684. Nutrients. 2018. PMID: 30400640 Free PMC article.
-
Impact of intrarectal chromofungin treatment on dendritic cells-related markers in different immune compartments in colonic inflammatory conditions.World J Gastroenterol. 2021 Dec 21;27(47):8138-8155. doi: 10.3748/wjg.v27.i47.8138. World J Gastroenterol. 2021. PMID: 35068859 Free PMC article.
-
Distinct roles for phosphoinositide 3-kinases γ and δ in malignant B cell migration.Leukemia. 2018 Sep;32(9):1958-1969. doi: 10.1038/s41375-018-0012-5. Epub 2018 Jan 31. Leukemia. 2018. PMID: 29479062 Free PMC article.
-
Semaphorin-3E attenuates intestinal inflammation through the regulation of the communication between splenic CD11C+ and CD4+ CD25- T-cells.Br J Pharmacol. 2019 May;176(9):1235-1250. doi: 10.1111/bph.14614. Epub 2019 Apr 1. Br J Pharmacol. 2019. PMID: 30736100 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

