The Many Roles of PCNA in Eukaryotic DNA Replication
- PMID: 27241932
- PMCID: PMC4890617
- DOI: 10.1016/bs.enz.2016.03.003
The Many Roles of PCNA in Eukaryotic DNA Replication
Abstract
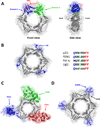
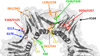
Proliferating cell nuclear antigen (PCNA) plays critical roles in many aspects of DNA replication and replication-associated processes, including translesion synthesis, error-free damage bypass, break-induced replication, mismatch repair, and chromatin assembly. Since its discovery, our view of PCNA has evolved from a replication accessory factor to the hub protein in a large protein-protein interaction network that organizes and orchestrates many of the key events at the replication fork. We begin this review article with an overview of the structure and function of PCNA. We discuss the ways its many interacting partners bind and how these interactions are regulated by posttranslational modifications such as ubiquitylation and sumoylation. We then explore the many roles of PCNA in normal DNA replication and in replication-coupled DNA damage tolerance and repair processes. We conclude by considering how PCNA can interact physically with so many binding partners to carry out its numerous roles. We propose that there is a large, dynamic network of linked PCNA molecules at and around the replication fork. This network would serve to increase the local concentration of all the proteins necessary for DNA replication and replication-associated processes and to regulate their various activities.
Keywords: Break-induced replication; DNA polymerase; DNA repair; DNA replication; Mismatch repair; PCNA; Processivity factor; Proliferating cell nuclear antigen; Sliding clamp; Translesion synthesis.
© 2016 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Eukaryotic translesion synthesis: Choosing the right tool for the job.DNA Repair (Amst). 2018 Nov;71:127-134. doi: 10.1016/j.dnarep.2018.08.016. Epub 2018 Aug 24. DNA Repair (Amst). 2018. PMID: 30174299 Free PMC article. Review.
-
Ub-family modifications at the replication fork: Regulating PCNA-interacting components.FEBS Lett. 2011 Sep 16;585(18):2920-8. doi: 10.1016/j.febslet.2011.08.008. Epub 2011 Aug 12. FEBS Lett. 2011. PMID: 21846465 Review.
-
Replication protein A dynamically regulates monoubiquitination of proliferating cell nuclear antigen.J Biol Chem. 2019 Mar 29;294(13):5157-5168. doi: 10.1074/jbc.RA118.005297. Epub 2019 Jan 30. J Biol Chem. 2019. PMID: 30700555 Free PMC article.
-
Functions of Multiple Clamp and Clamp-Loader Complexes in Eukaryotic DNA Replication.Adv Exp Med Biol. 2017;1042:135-162. doi: 10.1007/978-981-10-6955-0_7. Adv Exp Med Biol. 2017. PMID: 29357057 Review.
-
Control of DNA Damage Bypass by Ubiquitylation of PCNA.Genes (Basel). 2020 Jan 29;11(2):138. doi: 10.3390/genes11020138. Genes (Basel). 2020. PMID: 32013080 Free PMC article. Review.
Cited by
-
Proliferating Cell Nuclear Antigen in the Era of Oncolytic Virotherapy.Viruses. 2024 Aug 7;16(8):1264. doi: 10.3390/v16081264. Viruses. 2024. PMID: 39205238 Free PMC article. Review.
-
The Induction of G2/M Phase Cell Cycle Arrest and Apoptosis by the Chalcone Derivative 1C in Sensitive and Resistant Ovarian Cancer Cells Is Associated with ROS Generation.Int J Mol Sci. 2024 Jul 9;25(14):7541. doi: 10.3390/ijms25147541. Int J Mol Sci. 2024. PMID: 39062784 Free PMC article.
-
PARP10 promotes the repair of nascent strand DNA gaps through RAD18 mediated translesion synthesis.Nat Commun. 2024 Jul 23;15(1):6197. doi: 10.1038/s41467-024-50429-3. Nat Commun. 2024. PMID: 39043663 Free PMC article.
-
RNF212B E3 ligase is essential for crossover designation and maturation during male and female meiosis in the mouse.Proc Natl Acad Sci U S A. 2024 Jun 18;121(25):e2320995121. doi: 10.1073/pnas.2320995121. Epub 2024 Jun 12. Proc Natl Acad Sci U S A. 2024. PMID: 38865271 Free PMC article.
-
The bacterial DNA sliding clamp, β-clamp: structure, interactions, dynamics and drug discovery.Cell Mol Life Sci. 2024 May 30;81(1):245. doi: 10.1007/s00018-024-05252-w. Cell Mol Life Sci. 2024. PMID: 38814467 Free PMC article. Review.
References
-
- Miyachi K, Fritzler MJ, Tan EM. Autoantibody to a nuclear antigen in proliferating cells. J Immunol. 1978;121(6):2228–2234. - PubMed
-
- Bravo R, et al. Identification of a nuclear and of a cytoplasmic polypeptide whose relative proportions are sensitive to changes in the rate of cell proliferation. Exp Cell Res. 1981;136(2):311–319. - PubMed
-
- Mathews MB, et al. Identity of the proliferating cell nuclear antigen and cyclin. Nature. 1984;309(5966):374–376. - PubMed
-
- Madsen P, Celis JE. S-phase patterns of cyclin (PCNA) antigen staining resemble topographical patterns of DNA synthesis. A role for cyclin in DNA replication? FEBS Lett. 1985;193(1):5–11. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
