The Hemogenic Competence of Endothelial Progenitors Is Restricted by Runx1 Silencing during Embryonic Development
- PMID: 27239041
- PMCID: PMC4906370
- DOI: 10.1016/j.celrep.2016.05.001
The Hemogenic Competence of Endothelial Progenitors Is Restricted by Runx1 Silencing during Embryonic Development
Abstract
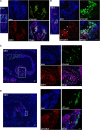
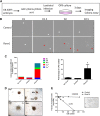
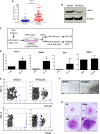
It is now well-established that hematopoietic stem cells (HSCs) and progenitor cells originate from a specialized subset of endothelium, termed hemogenic endothelium (HE), via an endothelial-to-hematopoietic transition. However, the molecular mechanisms determining which endothelial progenitors possess this hemogenic potential are currently unknown. Here, we investigated the changes in hemogenic potential in endothelial progenitors at the early stages of embryonic development. Using an ETV2::GFP reporter mouse to isolate emerging endothelial progenitors, we observed a dramatic decrease in hemogenic potential between embryonic day (E)7.5 and E8.5. At the molecular level, Runx1 is expressed at much lower levels in E8.5 intra-embryonic progenitors, while Bmi1 expression is increased. Remarkably, the ectopic expression of Runx1 in these progenitors fully restores their hemogenic potential, as does the suppression of BMI1 function. Altogether, our data demonstrate that hemogenic competency in recently specified endothelial progenitors is restrained through the active silencing of Runx1 expression.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
The Role of Runx1 in Embryonic Blood Cell Formation.Adv Exp Med Biol. 2017;962:47-64. doi: 10.1007/978-981-10-3233-2_4. Adv Exp Med Biol. 2017. PMID: 28299650 Free PMC article. Review.
-
ETV2 expression marks blood and endothelium precursors, including hemogenic endothelium, at the onset of blood development.Dev Dyn. 2012 Sep;241(9):1454-64. doi: 10.1002/dvdy.23825. Epub 2012 Jul 16. Dev Dyn. 2012. PMID: 22733530
-
Tuning apicobasal polarity and junctional recycling in the hemogenic endothelium orchestrates the morphodynamic complexity of emerging pre-hematopoietic stem cells.Elife. 2024 May 29;12:RP91429. doi: 10.7554/eLife.91429. Elife. 2024. PMID: 38809590 Free PMC article.
-
Runx1 is sufficient for blood cell formation from non-hemogenic endothelial cells in vivo only during early embryogenesis.Development. 2018 Jan 29;145(2):dev158162. doi: 10.1242/dev.158162. Development. 2018. PMID: 29361566 Free PMC article.
-
Specification and function of hemogenic endothelium during embryogenesis.Cell Mol Life Sci. 2016 Apr;73(8):1547-67. doi: 10.1007/s00018-016-2134-0. Epub 2016 Feb 5. Cell Mol Life Sci. 2016. PMID: 26849156 Free PMC article. Review.
Cited by
-
The T-box transcription factor Eomesodermin governs haemogenic competence of yolk sac mesodermal progenitors.Nat Cell Biol. 2021 Jan;23(1):61-74. doi: 10.1038/s41556-020-00611-8. Epub 2021 Jan 8. Nat Cell Biol. 2021. PMID: 33420489 Free PMC article.
-
Lineage-tracing hematopoietic stem cell origins in vivo to efficiently make human HLF+ HOXA+ hematopoietic progenitors from pluripotent stem cells.Dev Cell. 2024 May 6;59(9):1110-1131.e22. doi: 10.1016/j.devcel.2024.03.003. Epub 2024 Apr 2. Dev Cell. 2024. PMID: 38569552 Free PMC article.
-
The Role of Runx1 in Embryonic Blood Cell Formation.Adv Exp Med Biol. 2017;962:47-64. doi: 10.1007/978-981-10-3233-2_4. Adv Exp Med Biol. 2017. PMID: 28299650 Free PMC article. Review.
-
CHD7 and Runx1 interaction provides a braking mechanism for hematopoietic differentiation.Proc Natl Acad Sci U S A. 2020 Sep 22;117(38):23626-23635. doi: 10.1073/pnas.2003228117. Epub 2020 Sep 3. Proc Natl Acad Sci U S A. 2020. PMID: 32883883 Free PMC article.
-
Transcriptional control of blood cell emergence.FEBS Lett. 2019 Dec;593(23):3304-3315. doi: 10.1002/1873-3468.13585. Epub 2019 Aug 31. FEBS Lett. 2019. PMID: 31432499 Free PMC article. Review.
References
-
- Baron M. Induction of embryonic hematopoietic and endothelial stem/progenitor cells by hedgehog-mediated signals. Differentiation. 2001;68:175–185. - PubMed
-
- Boisset J.C., van Cappellen W., Andrieu-Soler C., Galjart N., Dzierzak E., Robin C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–120. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

