Promotion of RAD51-Mediated Homologous DNA Pairing by the RAD51AP1-UAF1 Complex
- PMID: 27239033
- PMCID: PMC5381662
- DOI: 10.1016/j.celrep.2016.05.007
Promotion of RAD51-Mediated Homologous DNA Pairing by the RAD51AP1-UAF1 Complex
Abstract
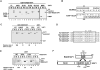
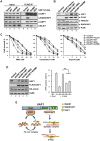
The UAF1-USP1 complex deubiquitinates FANCD2 during execution of the Fanconi anemia DNA damage response pathway. As such, UAF1 depletion results in persistent FANCD2 ubiquitination and DNA damage hypersensitivity. UAF1-deficient cells are also impaired for DNA repair by homologous recombination. Herein, we show that UAF1 binds DNA and forms a dimeric complex with RAD51AP1, an accessory factor of the RAD51 recombinase, and a trimeric complex with RAD51 through RAD51AP1. Two small ubiquitin-like modifier (SUMO)-like domains in UAF1 and a SUMO-interacting motif in RAD51AP1 mediate complex formation. Importantly, UAF1 enhances RAD51-mediated homologous DNA pairing in a manner that is dependent on complex formation with RAD51AP1 but independent of USP1. Mechanistically, RAD51AP1-UAF1 co-operates with RAD51 to assemble the synaptic complex, a critical nucleoprotein intermediate in homologous recombination, and cellular studies reveal the biological significance of the RAD51AP1-UAF1 protein complex. Our findings provide insights into an apparently USP1-independent role of UAF1 in genome maintenance.
Copyright © 2016 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures


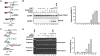
Similar articles
-
The USP1-UAF1 complex interacts with RAD51AP1 to promote homologous recombination repair.Cell Cycle. 2016 Oct;15(19):2636-2646. doi: 10.1080/15384101.2016.1209613. Epub 2016 Jul 27. Cell Cycle. 2016. PMID: 27463890 Free PMC article.
-
The DNA-binding activity of USP1-associated factor 1 is required for efficient RAD51-mediated homologous DNA pairing and homology-directed DNA repair.J Biol Chem. 2020 Jun 12;295(24):8186-8194. doi: 10.1074/jbc.RA120.013714. Epub 2020 Apr 29. J Biol Chem. 2020. PMID: 32350107 Free PMC article.
-
DNA requirement in FANCD2 deubiquitination by USP1-UAF1-RAD51AP1 in the Fanconi anemia DNA damage response.Nat Commun. 2019 Jun 28;10(1):2849. doi: 10.1038/s41467-019-10408-5. Nat Commun. 2019. PMID: 31253762 Free PMC article.
-
The Fanconi anemia ID2 complex: dueling saxes at the crossroads.Cell Cycle. 2014;13(19):2999-3015. doi: 10.4161/15384101.2014.956475. Cell Cycle. 2014. PMID: 25486561 Free PMC article. Review.
-
Role of RAD51AP1 in homologous recombination DNA repair and carcinogenesis.DNA Repair (Amst). 2017 Nov;59:76-81. doi: 10.1016/j.dnarep.2017.09.008. Epub 2017 Sep 22. DNA Repair (Amst). 2017. PMID: 28963981 Free PMC article. Review.
Cited by
-
Beyond reversal: ubiquitin and ubiquitin-like proteases and the orchestration of the DNA double strand break repair response.Biochem Soc Trans. 2019 Dec 20;47(6):1881-1893. doi: 10.1042/BST20190534. Biochem Soc Trans. 2019. PMID: 31769469 Free PMC article. Review.
-
The USP1-UAF1 complex interacts with RAD51AP1 to promote homologous recombination repair.Cell Cycle. 2016 Oct;15(19):2636-2646. doi: 10.1080/15384101.2016.1209613. Epub 2016 Jul 27. Cell Cycle. 2016. PMID: 27463890 Free PMC article.
-
A DUB-less step? Tighten up D-loop.Cell Cycle. 2016 Dec;15(23):3163-3164. doi: 10.1080/15384101.2016.1226603. Epub 2016 Sep 14. Cell Cycle. 2016. PMID: 27628628 Free PMC article. No abstract available.
-
Mechanistic Insights From Single-Molecule Studies of Repair of Double Strand Breaks.Front Cell Dev Biol. 2021 Nov 15;9:745311. doi: 10.3389/fcell.2021.745311. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34869333 Free PMC article. Review.
-
Walking a tightrope: The complex balancing act of R-loops in genome stability.Mol Cell. 2022 Jun 16;82(12):2267-2297. doi: 10.1016/j.molcel.2022.04.014. Epub 2022 May 3. Mol Cell. 2022. PMID: 35508167 Free PMC article. Review.
References
-
- Cohn Ma, Kowal P, Yang K, Haas W, Huang TT, Gygi SP, D'Andrea AD. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell. 2007;28:786–797. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

