RNA Interference Prevents Autosomal-Dominant Hearing Loss
- PMID: 27236922
- PMCID: PMC4908151
- DOI: 10.1016/j.ajhg.2016.03.028
RNA Interference Prevents Autosomal-Dominant Hearing Loss
Abstract
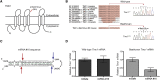
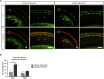
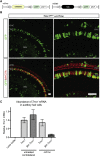
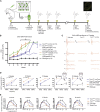
Hearing impairment is the most common sensory deficit. It is frequently caused by the expression of an allele carrying a single dominant missense mutation. Herein, we show that a single intracochlear injection of an artificial microRNA carried in a viral vector can slow progression of hearing loss for up to 35 weeks in the Beethoven mouse, a murine model of non-syndromic human deafness caused by a dominant gain-of-function mutation in Tmc1 (transmembrane channel-like 1). This outcome is noteworthy because it demonstrates the feasibility of RNA-interference-mediated suppression of an endogenous deafness-causing allele to slow progression of hearing loss. Given that most autosomal-dominant non-syndromic hearing loss in humans is caused by this mechanism of action, microRNA-based therapeutics might be broadly applicable as a therapy for this type of deafness.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Targeted Allele Suppression Prevents Progressive Hearing Loss in the Mature Murine Model of Human TMC1 Deafness.Mol Ther. 2019 Mar 6;27(3):681-690. doi: 10.1016/j.ymthe.2018.12.014. Epub 2019 Jan 7. Mol Ther. 2019. PMID: 30686588 Free PMC article.
-
Single and Dual Vector Gene Therapy with AAV9-PHP.B Rescues Hearing in Tmc1 Mutant Mice.Mol Ther. 2021 Mar 3;29(3):973-988. doi: 10.1016/j.ymthe.2020.11.016. Epub 2020 Nov 17. Mol Ther. 2021. PMID: 33212302 Free PMC article.
-
Mutation-agnostic RNA interference with engineered replacement rescues Tmc1-related hearing loss.Life Sci Alliance. 2022 Dec 27;6(3):e202201592. doi: 10.26508/lsa.202201592. Print 2023 Mar. Life Sci Alliance. 2022. PMID: 36574989 Free PMC article.
-
Genetics Of Human Hereditary Hearing Impairment.J Ayub Med Coll Abbottabad. 2017 Oct-Dec;29(4):671-676. J Ayub Med Coll Abbottabad. 2017. PMID: 29331002 Review.
-
Mouse tales from Kresge: the deafness mouse.J Am Acad Audiol. 2003 Aug;14(6):296-301. J Am Acad Audiol. 2003. PMID: 14552423 Review.
Cited by
-
Hearing Loss: Genetic Testing, Current Advances and the Situation in Latin America.Genes (Basel). 2024 Jan 29;15(2):178. doi: 10.3390/genes15020178. Genes (Basel). 2024. PMID: 38397168 Free PMC article. Review.
-
CRISPR/Cas9: targeted genome editing for the treatment of hereditary hearing loss.J Appl Genet. 2020 Feb;61(1):51-65. doi: 10.1007/s13353-019-00535-6. Epub 2020 Jan 7. J Appl Genet. 2020. PMID: 31912450 Review.
-
Improved TMC1 gene therapy restores hearing and balance in mice with genetic inner ear disorders.Nat Commun. 2019 Jan 22;10(1):236. doi: 10.1038/s41467-018-08264-w. Nat Commun. 2019. PMID: 30670701 Free PMC article.
-
Genome and base editing for genetic hearing loss.Hear Res. 2020 Sep 1;394:107958. doi: 10.1016/j.heares.2020.107958. Epub 2020 Apr 5. Hear Res. 2020. PMID: 32334889 Free PMC article. Review.
-
Gene editing in a Myo6 semi-dominant mouse model rescues auditory function.Mol Ther. 2022 Jan 5;30(1):105-118. doi: 10.1016/j.ymthe.2021.06.015. Epub 2021 Jun 24. Mol Ther. 2022. PMID: 34174443 Free PMC article.
References
-
- Smith R.J., Bale J.F., Jr., White K.R. Sensorineural hearing loss in children. Lancet. 2005;365:879–890. - PubMed
-
- Maeda Y., Fukushima K., Nishizaki K., Smith R.J. In vitro and in vivo suppression of GJB2 expression by RNA interference. Hum. Mol. Genet. 2005;14:1641–1650. - PubMed
-
- Maeda Y., Fukushima K., Kawasaki A., Nishizaki K., Smith R.J. Cochlear expression of a dominant-negative GJB2R75W construct delivered through the round window membrane in mice. Neurosci. Res. 2007;58:250–254. - PubMed
-
- Vreugde S., Erven A., Kros C.J., Marcotti W., Fuchs H., Kurima K., Wilcox E.R., Friedman T.B., Griffith A.J., Balling R. Beethoven, a mouse model for dominant, progressive hearing loss DFNA36. Nat. Genet. 2002;30:257–258. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

