The Philadelphia chromosome in leukemogenesis
- PMID: 27233483
- PMCID: PMC4896164
- DOI: 10.1186/s40880-016-0108-0
The Philadelphia chromosome in leukemogenesis
Abstract
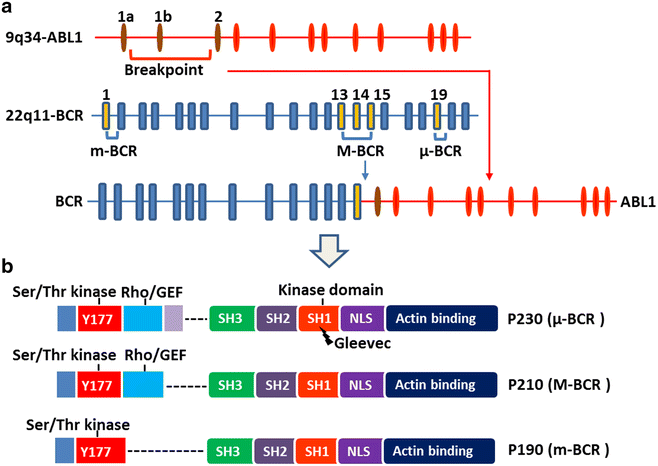
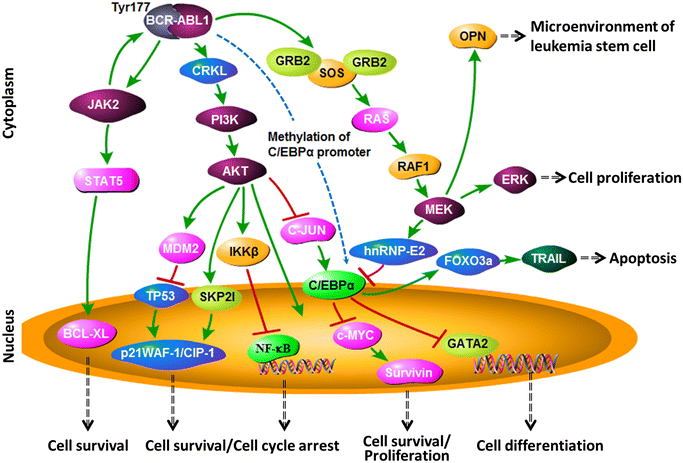
The truncated chromosome 22 that results from the reciprocal translocation t(9;22)(q34;q11) is known as the Philadelphia chromosome (Ph) and is a hallmark of chronic myeloid leukemia (CML). In leukemia cells, Ph not only impairs the physiological signaling pathways but also disrupts genomic stability. This aberrant fusion gene encodes the breakpoint cluster region-proto-oncogene tyrosine-protein kinase (BCR-ABL1) oncogenic protein with persistently enhanced tyrosine kinase activity. The kinase activity is responsible for maintaining proliferation, inhibiting differentiation, and conferring resistance to cell death. During the progression of CML from the chronic phase to the accelerated phase and then to the blast phase, the expression patterns of different BCR-ABL1 transcripts vary. Each BCR-ABL1 transcript is present in a distinct leukemia phenotype, which predicts both response to therapy and clinical outcome. Besides CML, the Ph is found in acute lymphoblastic leukemia, acute myeloid leukemia, and mixed-phenotype acute leukemia. Here, we provide an overview of the clinical presentation and cellular biology of different phenotypes of Ph-positive leukemia and highlight key findings regarding leukemogenesis.
Keywords: BCR-ABL1; Chronic myeloid leukemia; Philadelphia chromosome; Signaling pathway; Translocations.
Figures
Similar articles
-
Leukemia cell lines: in vitro models for the study of Philadelphia chromosome-positive leukemia.Leuk Res. 1999 Mar;23(3):207-15. doi: 10.1016/s0145-2126(98)00171-4. Leuk Res. 1999. PMID: 10071072 Review.
-
Overview of clinical and genetic features of CML patients with variant Philadelphia translocations involving chromosome 7: A case series.Leuk Res. 2021 Dec;111:106725. doi: 10.1016/j.leukres.2021.106725. Epub 2021 Oct 4. Leuk Res. 2021. PMID: 34634595
-
Unusual expression of mRNA typical of Philadelphia positive acute lymphoblastic leukemia detected in chronic myeloid leukemia.Am J Hematol. 1996 Jul;52(3):129-34. doi: 10.1002/(SICI)1096-8652(199607)52:3<129::AID-AJH1>3.0.CO;2-V. Am J Hematol. 1996. PMID: 8756076
-
Ponatinib in the treatment of chronic myeloid leukemia and philadelphia chromosome positive acute lymphoblastic leukemia.Future Oncol. 2019 Jan;15(3):257-269. doi: 10.2217/fon-2018-0371. Epub 2018 Sep 25. Future Oncol. 2019. PMID: 30251548 Review.
-
A rare BCR-ABL1 transcript in Philadelphia-positive acute myeloid leukemia: case report and literature review.BMC Cancer. 2019 Jan 10;19(1):50. doi: 10.1186/s12885-019-5265-5. BMC Cancer. 2019. PMID: 30630459 Free PMC article. Review.
Cited by
-
Genion, an accurate tool to detect gene fusion from long transcriptomics reads.BMC Genomics. 2022 Feb 14;23(1):129. doi: 10.1186/s12864-022-08339-5. BMC Genomics. 2022. PMID: 35164688 Free PMC article.
-
Molecular and Clinical Insights in the Increasing Detection of BCR::ABL1 p190+ in Adult Acute Myeloid Leukemia Patients.In Vivo. 2024 Jul-Aug;38(4):2016-2023. doi: 10.21873/invivo.13659. In Vivo. 2024. PMID: 38936913 Free PMC article.
-
Transcriptional activation of the miR-17-92 cluster is involved in the growth-promoting effects of MYB in human Ph-positive leukemia cells.Haematologica. 2019 Jan;104(1):82-92. doi: 10.3324/haematol.2018.191213. Epub 2018 Aug 3. Haematologica. 2019. PMID: 30076175 Free PMC article.
-
Historical Perspective and Current Trends in Anticancer Drug Development.Cancers (Basel). 2024 May 15;16(10):1878. doi: 10.3390/cancers16101878. Cancers (Basel). 2024. PMID: 38791957 Free PMC article. Review.
-
VNTR polymorphism in the breakpoint region of ABL1 and susceptibility to bladder cancer.BMC Med Genomics. 2021 May 5;14(1):121. doi: 10.1186/s12920-021-00968-1. BMC Med Genomics. 2021. PMID: 33952249 Free PMC article.
References
-
- Nowell PC. A minute chromosome in human chronic granulocytic leukemia. Science. 1960;132:497–501.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

