Interferon regulatory factor 4 attenuates Notch signaling to suppress the development of chronic lymphocytic leukemia
- PMID: 27232759
- PMCID: PMC5173044
- DOI: 10.18632/oncotarget.9596
Interferon regulatory factor 4 attenuates Notch signaling to suppress the development of chronic lymphocytic leukemia
Abstract
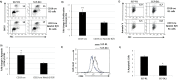
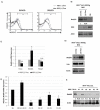
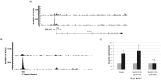
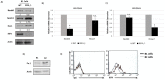
Molecular pathogenesis of Chronic Lymphocytic Leukemia (CLL) is not fully elucidated. Genome wide association studies have linked Interferon Regulatory Factor 4 (IRF4) to the development of CLL. We recently established a causal relationship between low levels of IRF4 and development of CLL. However, the molecular mechanism through which IRF4 suppresses CLL development remains unclear. Deregulation of Notch signaling pathway has been identified as one of the most recurrent molecular anomalies in the pathogenesis of CLL. Yet, the role of Notch signaling as well as its regulation during CLL development remains poorly understood. Previously, we demonstrated that IRF4 deficient mice expressing immunoglobulin heavy chain Vh11 (IRF4-/-Vh11) developed spontaneous CLL with complete penetrance. In this study, we show that elevated Notch2 expression and the resulting hyperactivation of Notch signaling are common features of IRF4-/-Vh11 CLL cells. Our studies further reveal that Notch signaling is indispensable for CLL development in the IRF4-/-Vh11 mice. Moreover, we identify E3 ubiquitin ligase Nedd4, which targets Notch for degradation, as a direct target of IRF4 in CLL cells and their precursors. Collectively, our studies provide the first in vivo evidence for an essential role of Notch signaling in the development of CLL and establish IRF4 as a critical regulator of Notch signaling during CLL development.
Keywords: B1 cells; CLL; IRF4; Notch signaling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
A role for IRF4 in the development of CLL.Blood. 2013 Oct 17;122(16):2848-55. doi: 10.1182/blood-2013-03-492769. Epub 2013 Aug 7. Blood. 2013. PMID: 23926303 Free PMC article.
-
Accelerated development of chronic lymphocytic leukemia in New Zealand Black mice expressing a low level of interferon regulatory factor 4.J Biol Chem. 2013 Sep 13;288(37):26430-40. doi: 10.1074/jbc.M113.475913. Epub 2013 Jul 29. J Biol Chem. 2013. PMID: 23897826 Free PMC article.
-
Activation of Notch and Myc Signaling via B-cell-Restricted Depletion of Dnmt3a Generates a Consistent Murine Model of Chronic Lymphocytic Leukemia.Cancer Res. 2021 Dec 15;81(24):6117-6130. doi: 10.1158/0008-5472.CAN-21-1273. Epub 2021 Oct 22. Cancer Res. 2021. PMID: 34686499 Free PMC article.
-
[Notch signal pathway and chronic lymphocytic leukemia].Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2014 Oct;22(5):1472-5. doi: 10.7534/j.issn.1009-2137.2014.05.055. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2014. PMID: 25338610 Review. Chinese.
-
p53 and Notch signaling in chronic lymphocytic leukemia: clues to identifying novel therapeutic strategies.Leukemia. 2011 Sep;25(9):1400-7. doi: 10.1038/leu.2011.103. Epub 2011 May 13. Leukemia. 2011. PMID: 21566651 Review.
Cited by
-
Mouse Models in the Study of Mature B-Cell Malignancies.Cold Spring Harb Perspect Med. 2021 Apr 1;11(4):a034827. doi: 10.1101/cshperspect.a034827. Cold Spring Harb Perspect Med. 2021. PMID: 32398289 Free PMC article. Review.
-
Interferon regulatory factor 4 (IRF4) is overexpressed in human non‑small cell lung cancer (NSCLC) and activates the Notch signaling pathway.Mol Med Rep. 2017 Nov;16(5):6034-6040. doi: 10.3892/mmr.2017.7319. Epub 2017 Aug 22. Mol Med Rep. 2017. PMID: 28849037 Free PMC article.
-
Emerging roles of the HECT-type E3 ubiquitin ligases in hematological malignancies.Discov Oncol. 2021 Oct 8;12(1):39. doi: 10.1007/s12672-021-00435-4. Discov Oncol. 2021. PMID: 35201500 Free PMC article. Review.
-
Chronic lymphocytic leukaemia.Nat Rev Dis Primers. 2017 Jan 19;3:16096. doi: 10.1038/nrdp.2016.96. Nat Rev Dis Primers. 2017. PMID: 28102226 Free PMC article. Review.
-
NOTCH1-mutated chronic lymphocytic leukemia cells are characterized by a MYC-related overexpression of nucleophosmin 1 and ribosome-associated components.Leukemia. 2017 Nov;31(11):2407-2415. doi: 10.1038/leu.2017.90. Epub 2017 Mar 21. Leukemia. 2017. PMID: 28321119
References
-
- Fabbri G, Rasi S, Rossi D, Trifonov V, Khiabanian H, Ma J, Grunn A, Fangazio M, Capello D, Monti S, Cresta S, Gargiulo E, Forconi F, Guarini A, Arcaini L, Paulli M, et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. The Journal of experimental medicine. 2011;208:1389–1401. - PMC - PubMed
-
- Kiel MJ, Velusamy T, Betz BL, Zhao L, Weigelin HG, Chiang MY, Huebner-Chan DR, Bailey NG, Yang DT, Bhagat G, Miranda RN, Bahler DW, Medeiros LJ, Lim MS, Elenitoba-Johnson KS. Whole-genome sequencing identifies recurrent somatic NOTCH2 mutations in splenic marginal zone lymphoma. The Journal of experimental medicine. 2012;209:1553–1565. - PMC - PubMed
-
- Puente XS, Bea S, Valdes-Mas R, Villamor N, Gutierrez-Abril J, Martin-Subero JI, Munar M, Rubio-Perez C, Jares P, Aymerich M, Baumann T, Beekman R, Belver L, Carrio A, Castellano G, Clot G, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526:519–524. - PubMed
-
- Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N, Escaramis G, Jares P, Bea S, Gonzalez-Diaz M, Bassaganyas L, Baumann T, Juan M, Lopez-Guerra M, Colomer D, Tubio JM, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

