Structural and Functional Investigations of the Effector Protein LpiR1 from Legionella pneumophila
- PMID: 27226543
- PMCID: PMC4957058
- DOI: 10.1074/jbc.M115.708701
Structural and Functional Investigations of the Effector Protein LpiR1 from Legionella pneumophila
Abstract
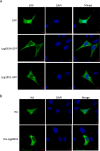
Legionella pneumophila is a causative agent of a severe pneumonia, known as Legionnaires' disease. Legionella pathogenicity is mediated by specific virulence factors, called bacterial effectors, which are injected into the invaded host cell by the bacterial type IV secretion system. Bacterial effectors are involved in complex interactions with the components of the host cell immune and signaling pathways, which eventually lead to bacterial survival and replication inside the mammalian cell. Structural and functional studies of bacterial effectors are, therefore, crucial for elucidating the mechanisms of Legionella virulence. Here we describe the crystal structure of the LpiR1 (Lpg0634) effector protein and investigate the effects of its overexpression in mammalian cells. LpiR1 is an α-helical protein that consists of two similar domains aligned in an antiparallel fashion. The hydrophilic cleft between the domains might serve as a binding site for a potential host cell interaction partner. LpiR1 binds the phosphate group at a conserved site and is stabilized by Mn(2+), Ca(2+), or Mg(2+) ions. When overexpressed in mammalian cells, a GFP-LpiR1 fusion protein is localized in the cytoplasm. Intracellular signaling antibody array analysis revealed small changes in the phosphorylation state of several components of the Akt signaling pathway in HEK293T cells overexpressing LpiR1.
Keywords: Legionella infection; bacterial pathogenesis; cell signaling; cellular localization; crystal structure; protein evolution; protein-protein interaction.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
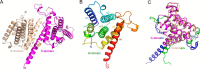



Similar articles
-
The Legionella effector RavD binds phosphatidylinositol-3-phosphate and helps suppress endolysosomal maturation of the Legionella-containing vacuole.J Biol Chem. 2019 Apr 19;294(16):6405-6415. doi: 10.1074/jbc.RA118.007086. Epub 2019 Feb 7. J Biol Chem. 2019. PMID: 30733336 Free PMC article.
-
The Sphingosine-1-Phosphate Lyase (LegS2) Contributes to the Restriction of Legionella pneumophila in Murine Macrophages.PLoS One. 2016 Jan 7;11(1):e0146410. doi: 10.1371/journal.pone.0146410. eCollection 2016. PLoS One. 2016. PMID: 26741365 Free PMC article.
-
Legionella pneumophila type IV effectors hijack the transcription and translation machinery of the host cell.Trends Cell Biol. 2014 Dec;24(12):771-8. doi: 10.1016/j.tcb.2014.06.002. Epub 2014 Jul 8. Trends Cell Biol. 2014. PMID: 25012125 Review.
-
Identification of two Legionella pneumophila effectors that manipulate host phospholipids biosynthesis.PLoS Pathog. 2012;8(11):e1002988. doi: 10.1371/journal.ppat.1002988. Epub 2012 Nov 1. PLoS Pathog. 2012. PMID: 23133385 Free PMC article.
-
Post-translational modifications of host proteins by Legionella pneumophila: a sophisticated survival strategy.Future Microbiol. 2012 Mar;7(3):369-81. doi: 10.2217/fmb.12.9. Future Microbiol. 2012. PMID: 22393890 Review.
Cited by
-
Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila.Mol Syst Biol. 2016 Dec 16;12(12):893. doi: 10.15252/msb.20167381. Mol Syst Biol. 2016. PMID: 27986836 Free PMC article.
-
Structural and functional study of Legionella pneumophila effector RavA.Protein Sci. 2021 May;30(5):940-955. doi: 10.1002/pro.4057. Epub 2021 Mar 9. Protein Sci. 2021. PMID: 33660322 Free PMC article.
References
-
- Vogel J. P., Andrews H. L., Wong S. K., and Isberg R. R. (1998) Conjugative transfer by the virulence system of Legionella pneumophila. Science 279, 873–876 - PubMed
-
- Vincent C. D., Friedman J. R., Jeong K. C., Buford E. C., Miller J. L., and Vogel J. P. (2006) Identification of the core transmembrane complex of the Legionella Dot/Icm type IV secretion system. Mol. Microbiol. 62, 1278–1291 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

