Mesoscale Modeling Reveals Hierarchical Looping of Chromatin Fibers Near Gene Regulatory Elements
- PMID: 27218881
- PMCID: PMC6268121
- DOI: 10.1021/acs.jpcb.6b03197
Mesoscale Modeling Reveals Hierarchical Looping of Chromatin Fibers Near Gene Regulatory Elements
Abstract
While it is well-recognized that chromatin loops play an important role in gene regulation, structural details regarding higher order chromatin loops are only emerging. Here we present a systematic study of restrained chromatin loops ranging from 25 to 427 nucleosomes (fibers of 5-80 Kb DNA in length), mimicking gene elements studied by 3C contact data. We find that hierarchical looping represents a stable configuration that can effectively bring distant regions of the GATA-4 gene together, satisfying connections reported by 3C experiments. Additionally, we find that restrained chromatin fibers larger than 100 nucleosomes (∼20Kb) form closed plectonemes, whereas fibers shorter than 100 nucleosomes form simple hairpin loops. By studying the dependence of loop structures on internal parameters, we show that loop features are sensitive to linker histone concentration, loop length, divalent ions, and DNA linker length. Specifically, increasing loop length, linker histone concentration, and divalent ion concentration are associated with increased persistence length (or decreased bending), while varying DNA linker length in a manner similar to experimentally observed "nucleosome free regions" (found near transcription start sites) disrupts intertwining and leads to loop opening and increased persistence length in linker histone depleted (-LH) fibers. Chromatin fiber structure sensitivity to these parameters, all of which vary throughout the cell cycle, tissue type, and species, suggests that caution is warranted when using uniform polymer models to fit chromatin conformation capture genome-wide data. Furthermore, the folding geometry we observe near the transcription initiation site of the GATA-4 gene suggests that hierarchical looping provides a structural mechanism for gene inhibition, and offers tunable parameters for design of gene regulation elements.
Figures
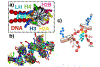


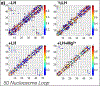

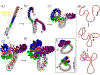
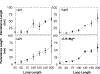

Similar articles
-
Linking Chromatin Fibers to Gene Folding by Hierarchical Looping.Biophys J. 2017 Feb 7;112(3):434-445. doi: 10.1016/j.bpj.2017.01.003. Epub 2017 Jan 31. Biophys J. 2017. PMID: 28153411 Free PMC article. Review.
-
Hierarchical looping of zigzag nucleosome chains in metaphase chromosomes.Proc Natl Acad Sci U S A. 2016 Feb 2;113(5):1238-43. doi: 10.1073/pnas.1518280113. Epub 2016 Jan 19. Proc Natl Acad Sci U S A. 2016. PMID: 26787893 Free PMC article.
-
Kilobase Pair Chromatin Fiber Contacts Promoted by Living-System-Like DNA Linker Length Distributions and Nucleosome Depletion.J Phys Chem B. 2017 Apr 20;121(15):3882-3894. doi: 10.1021/acs.jpcb.7b00998. Epub 2017 Mar 31. J Phys Chem B. 2017. PMID: 28299939 Free PMC article.
-
Chromatin Fiber Folding Directed by Cooperative Histone Tail Acetylation and Linker Histone Binding.Biophys J. 2018 May 22;114(10):2376-2385. doi: 10.1016/j.bpj.2018.03.008. Epub 2018 Apr 11. Biophys J. 2018. PMID: 29655483 Free PMC article.
-
Chromatin Higher-Order Folding: A Perspective with Linker DNA Angles.Biophys J. 2018 May 22;114(10):2290-2297. doi: 10.1016/j.bpj.2018.03.009. Epub 2018 Apr 6. Biophys J. 2018. PMID: 29628212 Free PMC article. Review.
Cited by
-
Mesoscale Modeling of Nucleosome-Binding Antibody PL2-6: Mono- versus Bivalent Chromatin Complexes.Biophys J. 2020 May 5;118(9):2066-2076. doi: 10.1016/j.bpj.2019.08.019. Epub 2019 Aug 22. Biophys J. 2020. PMID: 31668748 Free PMC article.
-
Large-scale simulations of nucleoprotein complexes: ribosomes, nucleosomes, chromatin, chromosomes and CRISPR.Curr Opin Struct Biol. 2019 Apr;55:104-113. doi: 10.1016/j.sbi.2019.03.004. Epub 2019 May 21. Curr Opin Struct Biol. 2019. PMID: 31125796 Free PMC article. Review.
-
Genome modeling: From chromatin fibers to genes.Curr Opin Struct Biol. 2023 Feb;78:102506. doi: 10.1016/j.sbi.2022.102506. Epub 2022 Dec 26. Curr Opin Struct Biol. 2023. PMID: 36577295 Free PMC article. Review.
-
Transferable model for chromosome architecture.Proc Natl Acad Sci U S A. 2016 Oct 25;113(43):12168-12173. doi: 10.1073/pnas.1613607113. Epub 2016 Sep 29. Proc Natl Acad Sci U S A. 2016. PMID: 27688758 Free PMC article.
-
Mesoscale modeling reveals formation of an epigenetically driven HOXC gene hub.Proc Natl Acad Sci U S A. 2019 Mar 12;116(11):4955-4962. doi: 10.1073/pnas.1816424116. Epub 2019 Feb 4. Proc Natl Acad Sci U S A. 2019. PMID: 30718394 Free PMC article.
References
-
- Worcel A; Benyajati C Higher order coiling of DNA in chromatin. Cell 1977, 12, 83–100. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

