Analysis of DNA gyrA Gene Mutation in Clinical and Environmental Ciprofloxacin-Resistant Isolates of Non-Tuberculous Mycobacteria Using Molecular Methods
- PMID: 27217921
- PMCID: PMC4870840
- DOI: 10.5812/jjm.30018
Analysis of DNA gyrA Gene Mutation in Clinical and Environmental Ciprofloxacin-Resistant Isolates of Non-Tuberculous Mycobacteria Using Molecular Methods
Abstract
Background: During the past several years, nontuberculous mycobacteria (NTM) have been reported as some of the most important agents of infection in immunocompromised patients.
Objectives: The aim of this study was to evaluate the ciprofloxacin susceptibility of clinical and environmental NTM species isolated from Isfahan province, Iran, using the agar dilution method, and to perform an analysis of gyrA gene-related ciprofloxacin resistance.
Materials and methods: A total of 41 clinical and environmental isolates of NTM were identified by conventional and multiplex PCR techniques. The isolates were separated out of water, blood, abscess, and bronchial samples. The susceptibility of the isolates to 1 µg/mL, 2 µg/mL and 4 µg/mL of ciprofloxacin concentrations was determined by the agar dilution method according to CLSI guidelines. A 120-bp area of the gyrA gene was amplified, and PCR-SSCP templates were defined using polyacrylamide gel electrophoresis. The 120-bp of gyrA amplicons with different PCR-SSCP patterns were sequenced.
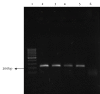
Results: The frequency of the identified isolates was as follows: Mycobacterium fortuitum, 27 cases; M. gordonae, 10 cases; M. smegmatis, one case; M. conceptionense, one case; and M. abscessus, two cases. All isolates except for M. abscessus were sensitive to all three concentrations of ciprofloxacin. The PCR-SSCP pattern of the gyrA gene of resistant M. abscessus isolates showed four different bands. The gyrA sequencing of resistant M. abscessus isolates showed 12 alterations in nucleotides compared to the M. abscessus ATCC 19977 resistant strain; however, the amino acid sequences were similar.
Conclusions: This study demonstrated the specificity and sensitivity of the PCR-SSCP method for finding mutations in the gyrA gene. Due to the sensitivity of most isolates to ciprofloxacin, this antibiotic should be considered an appropriate drug for the treatment of related diseases.
Keywords: Ciprofloxacin; DNA Gyrase A; PCR-SSCP.
Figures


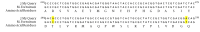

Similar articles
-
In Vitro Antimicrobial Susceptibility of Nontuberculous Mycobacteria in Iran.Microb Drug Resist. 2016 Mar;22(2):172-8. doi: 10.1089/mdr.2015.0134. Epub 2015 Oct 15. Microb Drug Resist. 2016. PMID: 26468990
-
Real time PCR for the rapid identification and drug susceptibility of Mycobacteria present in Bronchial washings.BMC Infect Dis. 2016 Oct 26;16(1):607. doi: 10.1186/s12879-016-1943-y. BMC Infect Dis. 2016. PMID: 27782812 Free PMC article.
-
[Detection of mutations in the gyrA and gyrB genes associated with fluoroquinolone-resistance among clinical isolates of Mycobacterium abscessus in China].Zhonghua Jie He He Hu Xi Za Zhi. 2015 Jul;38(7):507-10. Zhonghua Jie He He Hu Xi Za Zhi. 2015. PMID: 26703016 Chinese.
-
Evaluation of the new GenoType NTM-DR kit for the molecular detection of antimicrobial resistance in non-tuberculous mycobacteria.J Antimicrob Chemother. 2017 Jun 1;72(6):1669-1677. doi: 10.1093/jac/dkx021. J Antimicrob Chemother. 2017. PMID: 28333340
-
grlA and gyrA mutations and antimicrobial susceptibility in clinical isolates of ciprofloxacin- methicillin-resistant Staphylococcus aureus.Eur J Med Res. 2008 Aug 18;13(8):366-70. Eur J Med Res. 2008. PMID: 18952518
Cited by
-
Prevalence of nontuberculous mycobacteria and high efficacy of d-cycloserine and its synergistic effect with clarithromycin against Mycobacterium fortuitum and Mycobacterium abscessus.Infect Drug Resist. 2018 Dec 7;11:2521-2532. doi: 10.2147/IDR.S187554. eCollection 2018. Infect Drug Resist. 2018. PMID: 30573983 Free PMC article.
References
-
- Doucet-Populaire F, Buriankova K, Weiser J, Pernodet JL. Natural and acquired macrolide resistance in mycobacteria. Curr Drug Targets Infect Disord. 2002;2(4):355–70. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
