Enrichment of extracellular vesicles from tissues of the central nervous system by PROSPR
- PMID: 27216497
- PMCID: PMC4877958
- DOI: 10.1186/s13024-016-0108-1
Enrichment of extracellular vesicles from tissues of the central nervous system by PROSPR
Abstract
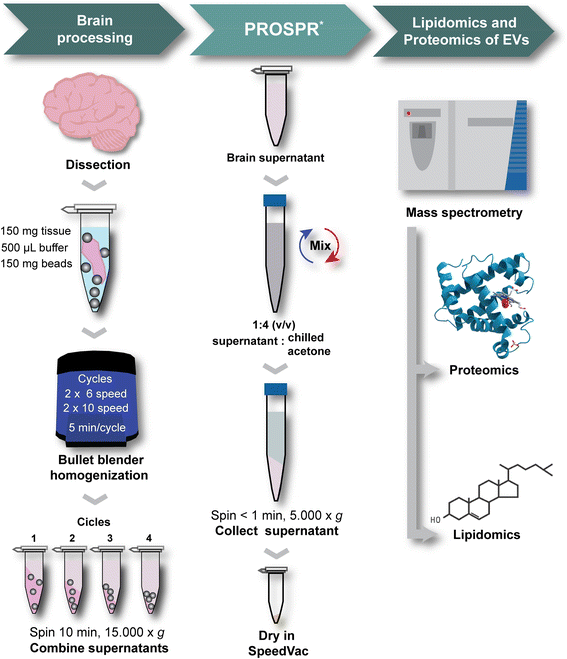
Background: Extracellular vesicles (EVs) act as key mediators of intercellular communication and are secreted and taken up by all cell types in the central nervous system (CNS). While detailed study of EV-based signaling is likely to significantly advance our understanding of human neurobiology, the technical challenges of isolating EVs from CNS tissues have limited their characterization using 'omics' technologies. We therefore developed a new Protein Organic Solvent Precipitation (PROSPR) method that can efficiently isolate the EV repertoire from human biological samples.
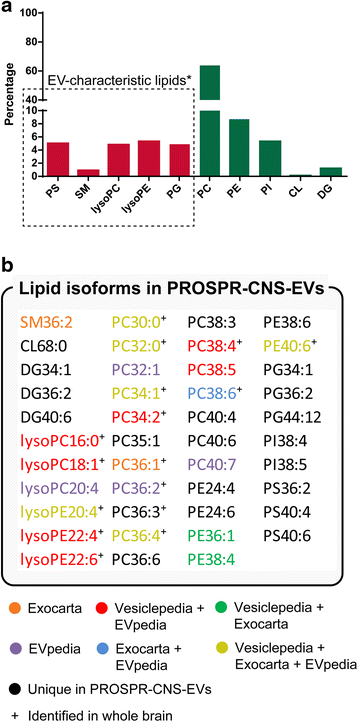
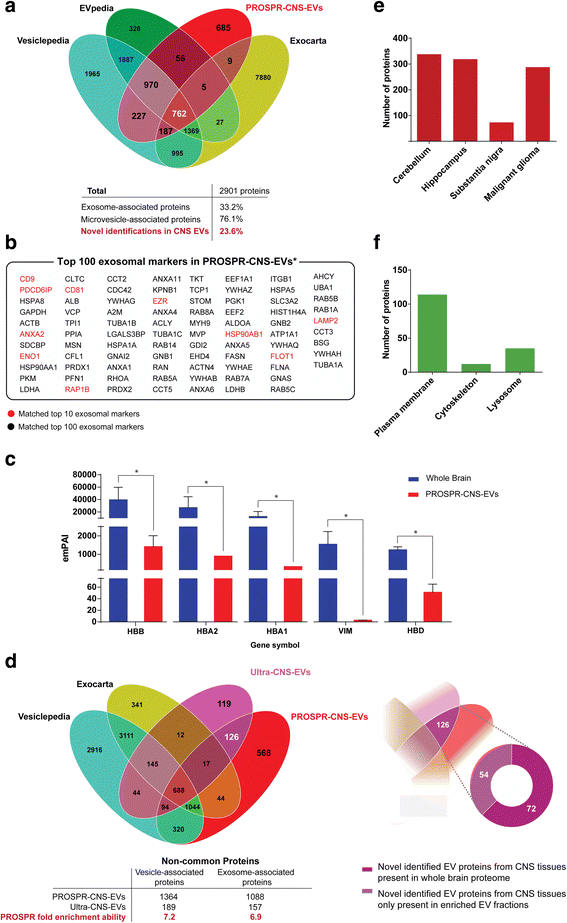
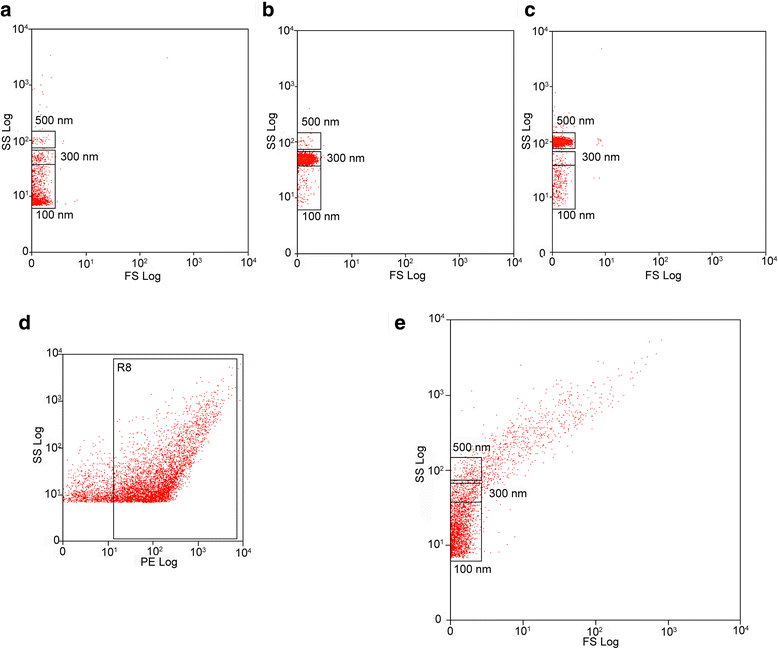
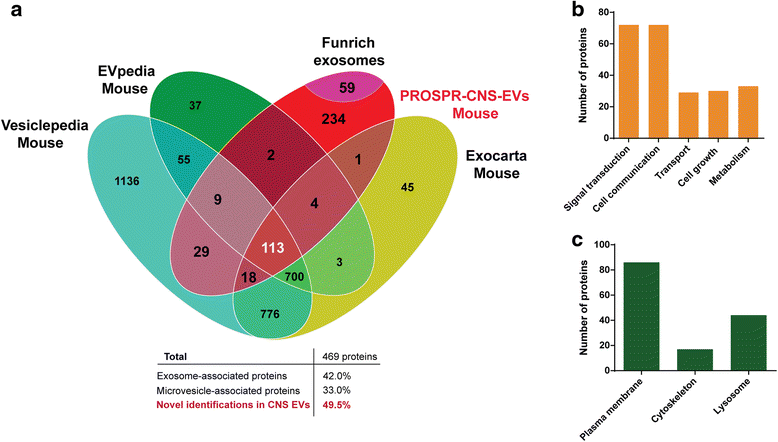
Results: In the current report, we present a novel experimental workflow that outlines the process of sample extraction and enrichment of CNS-derived EVs using PROSPR. Subsequent LC-MS/MS-based proteomic profiling of EVs enriched from brain homogenates successfully identified 86 of the top 100 exosomal markers. Proteomic profiling of PROSPR-enriched CNS EVs indicated that > 75 % of the proteins identified matched previously reported exosomal and microvesicle cargoes, while also expanded the known human EV-associated proteome with 685 novel identifications. Similarly, lipidomic characterization of enriched CNS vesicles not only identified previously reported EV-specific lipid families (PS, SM, lysoPC, lysoPE) but also uncovered novel lipid isoforms not previously detected in human EVs. Finally, dedicated flow cytometry of PROSPR-CNS-EVs revealed that ~80 % of total microparticles observed were exosomes ranging in diameter from ≤100 nm to 300 nm.
Conclusions: These data demonstrate that the optimized use of PROSPR represents an easy-to-perform and inexpensive method of enriching EVs from human CNS tissues for detailed characterization by 'omics' technologies. We predict that widespread use of the methodology described herein will greatly accelerate the study of EVs biology in neuroscience.
Keywords: Exosomes; Extracellular vesicles; Human brain; Lipidomics; Microvesicles; Proteomics; Tissue extraction.
Figures
Similar articles
-
Extracellular vesicles are rapidly purified from human plasma by PRotein Organic Solvent PRecipitation (PROSPR).Sci Rep. 2015 Sep 30;5:14664. doi: 10.1038/srep14664. Sci Rep. 2015. PMID: 26419333 Free PMC article.
-
Proteomic analysis of cerebrospinal fluid extracellular vesicles: a comprehensive dataset.J Proteomics. 2014 Jun 25;106:191-204. doi: 10.1016/j.jprot.2014.04.028. Epub 2014 Apr 24. J Proteomics. 2014. PMID: 24769233
-
Rapid Isolation of Extracellular Vesicles from Blood Plasma with Size-Exclusion Chromatography Followed by Mass Spectrometry-Based Proteomic Profiling.Methods Mol Biol. 2017;1660:295-302. doi: 10.1007/978-1-4939-7253-1_24. Methods Mol Biol. 2017. PMID: 28828666 Free PMC article.
-
Mass-spectrometry-based molecular characterization of extracellular vesicles: lipidomics and proteomics.J Proteome Res. 2015 Jun 5;14(6):2367-84. doi: 10.1021/pr501279t. Epub 2015 May 14. J Proteome Res. 2015. PMID: 25927954 Review.
-
Proteomic Analysis of Extracellular Vesicles for Cancer Diagnostics.Proteomics. 2019 Jan;19(1-2):e1800162. doi: 10.1002/pmic.201800162. Epub 2019 Jan 11. Proteomics. 2019. PMID: 30334355 Review.
Cited by
-
Toward a human brain extracellular vesicle atlas: Characteristics of extracellular vesicles from different brain regions, including small RNA and protein profiles.Interdiscip Med. 2023 Oct;1(4):e20230016. doi: 10.1002/INMD.20230016. Epub 2023 Aug 15. Interdiscip Med. 2023. PMID: 38089920 Free PMC article.
-
Therapeutic and Diagnostic Translation of Extracellular Vesicles in Cardiovascular Diseases: Roadmap to the Clinic.Circulation. 2021 Apr 6;143(14):1426-1449. doi: 10.1161/CIRCULATIONAHA.120.049254. Epub 2021 Apr 5. Circulation. 2021. PMID: 33819075 Free PMC article.
-
From biogenesis to aptasensors: advancements in analysis for tumor-derived extracellular vesicles research.Theranostics. 2024 Jul 2;14(10):4161-4183. doi: 10.7150/thno.95885. eCollection 2024. Theranostics. 2024. PMID: 38994022 Free PMC article. Review.
-
Research advances and challenges in tissue-derived extracellular vesicles.Front Mol Biosci. 2022 Dec 15;9:1036746. doi: 10.3389/fmolb.2022.1036746. eCollection 2022. Front Mol Biosci. 2022. PMID: 36589228 Free PMC article. Review.
-
Biogenesis, Isolation, and Detection of Exosomes and Their Potential in Therapeutics and Diagnostics.Biosensors (Basel). 2023 Aug 10;13(8):802. doi: 10.3390/bios13080802. Biosensors (Basel). 2023. PMID: 37622888 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

