SUMO5, a Novel Poly-SUMO Isoform, Regulates PML Nuclear Bodies
- PMID: 27211601
- PMCID: PMC4876461
- DOI: 10.1038/srep26509
SUMO5, a Novel Poly-SUMO Isoform, Regulates PML Nuclear Bodies
Abstract
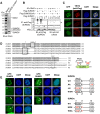
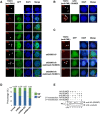
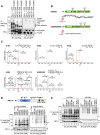
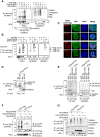
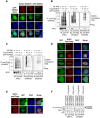
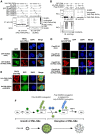
Promyelocytic leukemia nuclear bodies (PML-NBs) are PML-based nuclear structures that regulate various cellular processes. SUMOylation, the process of covalently conjugating small ubiquitin-like modifiers (SUMOs), is required for both the formation and the disruption of PML-NBs. However, detailed mechanisms of how SUMOylation regulates these processes remain unknown. Here we report that SUMO5, a novel SUMO variant, mediates the growth and disruption of PML-NBs. PolySUMO5 conjugation of PML at lysine 160 facilitates recruitment of PML-NB components, which enlarges PML-NBs. SUMO5 also increases polySUMO2/3 conjugation of PML, resulting in RNF4-mediated disruption of PML-NBs. The acute promyelocytic leukemia oncoprotein PML-RARα blocks SUMO5 conjugation of PML, causing cytoplasmic displacement of PML and disruption of PML-NBs. Our work not only identifies a new member of the SUMO family but also reveals the mechanistic basis of the PML-NB life cycle in human cells.
Figures

Similar articles
-
SUMOylation regulates the number and size of promyelocytic leukemia-nuclear bodies (PML-NBs) and arsenic perturbs SUMO dynamics on PML by insolubilizing PML in THP-1 cells.Arch Toxicol. 2022 Feb;96(2):545-558. doi: 10.1007/s00204-021-03195-w. Epub 2022 Jan 10. Arch Toxicol. 2022. PMID: 35001170
-
The promyelocytic leukemia protein isoform PML1 is an oncoprotein and a direct target of the antioxidant sulforaphane (SFN).Biochim Biophys Acta Mol Cell Res. 2020 Aug;1867(8):118707. doi: 10.1016/j.bbamcr.2020.118707. Epub 2020 Mar 31. Biochim Biophys Acta Mol Cell Res. 2020. PMID: 32243901
-
Respiratory syncytial virus induces NRF2 degradation through a promyelocytic leukemia protein - ring finger protein 4 dependent pathway.Free Radic Biol Med. 2017 Dec;113:494-504. doi: 10.1016/j.freeradbiomed.2017.10.380. Epub 2017 Oct 28. Free Radic Biol Med. 2017. PMID: 29107745 Free PMC article.
-
A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics.Int J Biol Sci. 2010 Jan 12;6(1):51-67. doi: 10.7150/ijbs.6.51. Int J Biol Sci. 2010. PMID: 20087442 Free PMC article. Review.
-
Unravelling the molecular interplay: SUMOylation, PML nuclear bodies and vascular cell activity in health and disease.Cell Signal. 2024 Jul;119:111156. doi: 10.1016/j.cellsig.2024.111156. Epub 2024 Apr 2. Cell Signal. 2024. PMID: 38574938 Review.
Cited by
-
The Four Homeostasis Knights: In Balance upon Post-Translational Modifications.Int J Mol Sci. 2022 Nov 21;23(22):14480. doi: 10.3390/ijms232214480. Int J Mol Sci. 2022. PMID: 36430960 Free PMC article. Review.
-
Role of Influenza A virus protein NS1 in regulating host nuclear body ND10 complex formation and its involvement in establishment of viral pathogenesis.PLoS One. 2024 Jan 2;19(1):e0295522. doi: 10.1371/journal.pone.0295522. eCollection 2024. PLoS One. 2024. PMID: 38166085 Free PMC article.
-
Histone sumoylation and chromatin dynamics.Nucleic Acids Res. 2021 Jun 21;49(11):6043-6052. doi: 10.1093/nar/gkab280. Nucleic Acids Res. 2021. PMID: 33885816 Free PMC article. Review.
-
SUMOylation in α-Synuclein Homeostasis and Pathology.Front Aging Neurosci. 2020 Jun 25;12:167. doi: 10.3389/fnagi.2020.00167. eCollection 2020. Front Aging Neurosci. 2020. PMID: 32670048 Free PMC article. Review.
-
Revisiting promyelocytic leukemia protein targeting by human cytomegalovirus immediate-early protein 1.PLoS Pathog. 2020 May 4;16(5):e1008537. doi: 10.1371/journal.ppat.1008537. eCollection 2020 May. PLoS Pathog. 2020. PMID: 32365141 Free PMC article.
References
-
- D’Orazi G. et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol 4, 11–19 (2002). - PubMed
-
- Szostecki C., Guldner H. H., Netter H. J. & Will H. Isolation and characterization of cDNA encoding a human nuclear antigen predominantly recognized by autoantibodies from patients with primary biliary cirrhosis. J Immunol 145, 4338–4347 (1990). - PubMed
-
- Skinner P. J. et al. Ataxin-1 with an expanded glutamine tract alters nuclear matrix-associated structures. Nature 389, 971–974 (1997). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

