TRAF3 signaling: Competitive binding and evolvability of adaptive viral molecular mimicry
- PMID: 27208423
- PMCID: PMC7117012
- DOI: 10.1016/j.bbagen.2016.05.021
TRAF3 signaling: Competitive binding and evolvability of adaptive viral molecular mimicry
Abstract
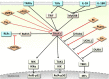
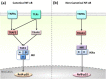
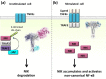
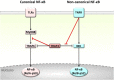
Background: The tumor necrosis factor receptor (TNFR) associated factor 3 (TRAF3) is a key node in innate and adaptive immune signaling pathways. TRAF3 negatively regulates the activation of the canonical and non-canonical NF-κB pathways and is one of the key proteins in antiviral immunity.
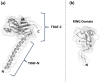
Scope of review: Here we provide a structural overview of TRAF3 signaling in terms of its competitive binding and consequences to the cellular network. For completion, we also include molecular mimicry of TRAF3 physiological partners by some viral proteins.
Major conclusions: By out-competing host partners, viral proteins aim to subvert TRAF3 antiviral action. Mechanistically, dynamic, competitive binding by the organism's own proteins and same-site adaptive pathogen mimicry follow the same conformational selection principles.
General significance: Our premise is that irrespective of the eliciting event - physiological or acquired pathogenic trait - pathway activation (or suppression) may embrace similar conformational principles. However, even though here we largely focus on competitive binding at a shared site, similar to physiological signaling other pathogen subversion mechanisms can also be at play. This article is part of a Special Issue entitled "System Genetics" Guest Editor: Dr. Yudong Cai and Dr. Tao Huang.
Keywords: Antiviral immunity; Cancer; Evolvable; Host-pathogen interactions; Inflammation; Structure.
Copyright © 2016 Elsevier B.V. All rights reserved.
Figures



Similar articles
-
Enterovirus D68 Protease 2Apro Targets TRAF3 To Subvert Host Innate Immune Responses.J Virol. 2021 Jan 13;95(3):e01856-20. doi: 10.1128/JVI.01856-20. Print 2021 Jan 13. J Virol. 2021. PMID: 33148796 Free PMC article.
-
The Nucleoprotein of H7N9 Influenza Virus Positively Regulates TRAF3-Mediated Innate Signaling and Attenuates Viral Virulence in Mice.J Virol. 2020 Nov 23;94(24):e01640-20. doi: 10.1128/JVI.01640-20. Print 2020 Nov 23. J Virol. 2020. PMID: 33028715 Free PMC article.
-
Differential requirements for tumor necrosis factor receptor-associated factor family proteins in CD40-mediated induction of NF-kappaB and Jun N-terminal kinase activation.J Biol Chem. 1999 Aug 6;274(32):22414-22. doi: 10.1074/jbc.274.32.22414. J Biol Chem. 1999. PMID: 10428814
-
Roles of tumor necrosis factor receptor associated factor 3 (TRAF3) and TRAF5 in immune cell functions.Immunol Rev. 2011 Nov;244(1):55-74. doi: 10.1111/j.1600-065X.2011.01055.x. Immunol Rev. 2011. PMID: 22017431 Free PMC article. Review.
-
Targeting signaling factors for degradation, an emerging mechanism for TRAF functions.Immunol Rev. 2015 Jul;266(1):56-71. doi: 10.1111/imr.12311. Immunol Rev. 2015. PMID: 26085207 Free PMC article. Review.
Cited by
-
Human papillomavirus and the landscape of secondary genetic alterations in oral cancers.Genome Res. 2019 Jan;29(1):1-17. doi: 10.1101/gr.241141.118. Epub 2018 Dec 18. Genome Res. 2019. PMID: 30563911 Free PMC article. Clinical Trial.
-
Dysregulated TRAF3 and BCL2 Expression Promotes Multiple Classes of Mature Non-hodgkin B Cell Lymphoma in Mice.Front Immunol. 2019 Jan 11;9:3114. doi: 10.3389/fimmu.2018.03114. eCollection 2018. Front Immunol. 2019. PMID: 30687320 Free PMC article.
-
Thrombocytopenia in COVID‑19 and vaccine‑induced thrombotic thrombocytopenia.Int J Mol Med. 2022 Mar;49(3):35. doi: 10.3892/ijmm.2022.5090. Epub 2022 Jan 21. Int J Mol Med. 2022. PMID: 35059730 Free PMC article.
-
Independent and core pathways in oncogenic KRAS signaling.Expert Rev Proteomics. 2016 Aug;13(8):711-6. doi: 10.1080/14789450.2016.1209417. Epub 2016 Jul 18. Expert Rev Proteomics. 2016. PMID: 27389825 Free PMC article. No abstract available.
-
TRAF Regulation of IL-17 Cytokine Signaling.Front Immunol. 2019 Jun 27;10:1293. doi: 10.3389/fimmu.2019.01293. eCollection 2019. Front Immunol. 2019. PMID: 31316496 Free PMC article. Review.
References
-
- Tsai C.J., Lin S.L., Wolfson H.J., Nussinov R. Protein-protein interfaces: architectures and interactions in protein-protein interfaces and in protein cores. Their similarities and differences. Crit. Rev. Biochem. Mol. Biol. 1996;31:127–152. - PubMed
-
- Keskin O., Ma B., Rogale K., Gunasekaran K., Nussinov R. Protein-protein interactions: organization, cooperativity and mapping in a bottom-up systems biology approach. Phys. Biol. 2005;2:S24–S35. - PubMed
-
- Akiva E., Babbitt P.C. Evolutionary reprograming of protein-protein interaction specificity. Cell. 2015;163:535–537. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

