Chromatibody, a novel non-invasive molecular tool to explore and manipulate chromatin in living cells
- PMID: 27206857
- PMCID: PMC4958301
- DOI: 10.1242/jcs.183103
Chromatibody, a novel non-invasive molecular tool to explore and manipulate chromatin in living cells
Abstract
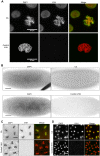
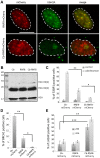
Chromatin function is involved in many cellular processes, its visualization or modification being essential in many developmental or cellular studies. Here, we present the characterization of chromatibody, a chromatin-binding single-domain, and explore its use in living cells. This non-intercalating tool specifically binds the heterodimer of H2A-H2B histones and displays a versatile reactivity, specifically labeling chromatin from yeast to mammals. We show that this genetically encoded probe, when fused to fluorescent proteins, allows non-invasive real-time chromatin imaging. Chromatibody is a dynamic chromatin probe that can be modulated. Finally, chromatibody is an efficient tool to target an enzymatic activity to the nucleosome, such as the DNA damage-dependent H2A ubiquitylation, which can modify this epigenetic mark at the scale of the genome and result in DNA damage signaling and repair defects. Taken together, these results identify chromatibody as a universal non-invasive tool for either in vivo chromatin imaging or to manipulate the chromatin landscape.
Keywords: Chromatin; Chromatin function; Epigenetic; Real-time imaging; Single-domain antibody.
© 2016. Published by The Company of Biologists Ltd.
Conflict of interest statement
D.J., J.V., B.S. and G.M. are co-inventors on the patent WO2014202745 A1, concerning the chromatibody discovery and applications. The other authors declare no conflict of interest.
Figures



Similar articles
-
DNA damage leaves its mark on chromatin.Cell Cycle. 2007 Oct 1;6(19):2355-9. doi: 10.4161/cc.6.19.4756. Epub 2007 Jul 18. Cell Cycle. 2007. PMID: 17703111
-
Histone H2A ubiquitination in transcriptional regulation and DNA damage repair.Int J Biochem Cell Biol. 2009 Jan;41(1):12-5. doi: 10.1016/j.biocel.2008.09.016. Epub 2008 Sep 26. Int J Biochem Cell Biol. 2009. PMID: 18929679 Review.
-
Histone H2A/H2B chaperones: from molecules to chromatin-based functions in plant growth and development.Plant J. 2015 Jul;83(1):78-95. doi: 10.1111/tpj.12830. Epub 2015 Apr 8. Plant J. 2015. PMID: 25781491 Review.
-
Transcription through chromatin: understanding a complex FACT.Biochim Biophys Acta. 2004 Mar 15;1677(1-3):87-99. doi: 10.1016/j.bbaexp.2003.09.017. Biochim Biophys Acta. 2004. PMID: 15020050 Review.
-
Open, repair and close again: chromatin dynamics and the response to UV-induced DNA damage.DNA Repair (Amst). 2011 Feb 7;10(2):119-25. doi: 10.1016/j.dnarep.2010.10.010. Epub 2010 Dec 3. DNA Repair (Amst). 2011. PMID: 21130713 Review.
Cited by
-
Functional Study of Haemophilus ducreyi Cytolethal Distending Toxin Subunit B.Toxins (Basel). 2020 Aug 19;12(9):530. doi: 10.3390/toxins12090530. Toxins (Basel). 2020. PMID: 32825080 Free PMC article.
-
Single-Domain Antibodies Represent Novel Alternatives to Monoclonal Antibodies as Targeting Agents against the Human Papillomavirus 16 E6 Protein.Int J Mol Sci. 2019 Apr 28;20(9):2088. doi: 10.3390/ijms20092088. Int J Mol Sci. 2019. PMID: 31035322 Free PMC article.
-
Nuclear lamina strain states revealed by intermolecular force biosensor.Nat Commun. 2023 Jun 30;14(1):3867. doi: 10.1038/s41467-023-39563-6. Nat Commun. 2023. PMID: 37391402 Free PMC article.
-
Chronic exposure to Cytolethal Distending Toxin (CDT) promotes a cGAS-dependent type I interferon response.Cell Mol Life Sci. 2021 Sep;78(17-18):6319-6335. doi: 10.1007/s00018-021-03902-x. Epub 2021 Jul 25. Cell Mol Life Sci. 2021. PMID: 34308492 Free PMC article.
-
Nanobodies Right in the Middle: Intrabodies as Toolbox to Visualize and Modulate Antigens in the Living Cell.Biomolecules. 2020 Dec 21;10(12):1701. doi: 10.3390/biom10121701. Biomolecules. 2020. PMID: 33371447 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

