Genome Therapy of Myotonic Dystrophy Type 1 iPS Cells for Development of Autologous Stem Cell Therapy
- PMID: 27203440
- PMCID: PMC5023370
- DOI: 10.1038/mt.2016.97
Genome Therapy of Myotonic Dystrophy Type 1 iPS Cells for Development of Autologous Stem Cell Therapy
Abstract
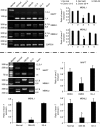
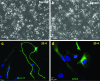
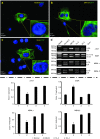
Myotonic dystrophy type 1 (DM1) is caused by expanded Cytosine-Thymine-Guanine (CTG) repeats in the 3'-untranslated region (3' UTR) of the Dystrophia myotonica protein kinase (DMPK) gene, for which there is no effective therapy. The objective of this study is to develop genome therapy in human DM1 induced pluripotent stem (iPS) cells to eliminate mutant transcripts and reverse the phenotypes for developing autologous stem cell therapy. The general approach involves targeted insertion of polyA signals (PASs) upstream of DMPK CTG repeats, which will lead to premature termination of transcription and elimination of toxic mutant transcripts. Insertion of PASs was mediated by homologous recombination triggered by site-specific transcription activator-like effector nuclease (TALEN)-induced double-strand break. We found genome-treated DM1 iPS cells continue to maintain pluripotency. The insertion of PASs led to elimination of mutant transcripts and complete disappearance of nuclear RNA foci and reversal of aberrant splicing in linear-differentiated neural stem cells, cardiomyocytes, and teratoma tissues. In conclusion, genome therapy by insertion of PASs upstream of the expanded DMPK CTG repeats prevented the production of toxic mutant transcripts and reversal of phenotypes in DM1 iPS cells and their progeny. These genetically-treated iPS cells will have broad clinical application in developing autologous stem cell therapy for DM1.
Figures
Similar articles
-
Genome modification leads to phenotype reversal in human myotonic dystrophy type 1 induced pluripotent stem cell-derived neural stem cells.Stem Cells. 2015 Jun;33(6):1829-38. doi: 10.1002/stem.1970. Stem Cells. 2015. PMID: 25702800 Free PMC article.
-
Therapeutic Genome Editing for Myotonic Dystrophy Type 1 Using CRISPR/Cas9.Mol Ther. 2018 Nov 7;26(11):2617-2630. doi: 10.1016/j.ymthe.2018.09.003. Epub 2018 Sep 11. Mol Ther. 2018. PMID: 30274788 Free PMC article.
-
Genome Editing of Expanded CTG Repeats within the Human DMPK Gene Reduces Nuclear RNA Foci in the Muscle of DM1 Mice.Mol Ther. 2019 Aug 7;27(8):1372-1388. doi: 10.1016/j.ymthe.2019.05.021. Epub 2019 Jun 5. Mol Ther. 2019. PMID: 31253581 Free PMC article.
-
Application of CRISPR-Cas9-Mediated Genome Editing for the Treatment of Myotonic Dystrophy Type 1.Mol Ther. 2020 Dec 2;28(12):2527-2539. doi: 10.1016/j.ymthe.2020.10.005. Epub 2020 Oct 14. Mol Ther. 2020. PMID: 33171139 Free PMC article. Review.
-
Myotonic dystrophy type 1: role of CCG, CTC and CGG interruptions within DMPK alleles in the pathogenesis and molecular diagnosis.Clin Genet. 2017 Oct;92(4):355-364. doi: 10.1111/cge.12954. Epub 2017 Feb 22. Clin Genet. 2017. PMID: 27991661 Review.
Cited by
-
Unexpected Mutations by CRISPR-Cas9 CTG Repeat Excision in Myotonic Dystrophy and Use of CRISPR Interference as an Alternative Approach.Mol Ther Methods Clin Dev. 2020 May 22;18:131-144. doi: 10.1016/j.omtm.2020.05.024. eCollection 2020 Sep 11. Mol Ther Methods Clin Dev. 2020. PMID: 32637445 Free PMC article.
-
Daunorubicin reduces MBNL1 sequestration caused by CUG-repeat expansion and rescues cardiac dysfunctions in a Drosophila model of myotonic dystrophy.Dis Model Mech. 2018 Apr 23;11(4):dmm032557. doi: 10.1242/dmm.032557. Dis Model Mech. 2018. PMID: 29592894 Free PMC article.
-
Myotonic Dystrophy and Developmental Regulation of RNA Processing.Compr Physiol. 2018 Mar 25;8(2):509-553. doi: 10.1002/cphy.c170002. Compr Physiol. 2018. PMID: 29687899 Free PMC article. Review.
-
Modelling inherited cardiac disease using human induced pluripotent stem cell-derived cardiomyocytes: progress, pitfalls, and potential.Cardiovasc Res. 2018 Dec 1;114(14):1828-1842. doi: 10.1093/cvr/cvy208. Cardiovasc Res. 2018. PMID: 30169602 Free PMC article. Review.
-
Furamidine Rescues Myotonic Dystrophy Type I Associated Mis-Splicing through Multiple Mechanisms.ACS Chem Biol. 2018 Sep 21;13(9):2708-2718. doi: 10.1021/acschembio.8b00646. Epub 2018 Aug 27. ACS Chem Biol. 2018. PMID: 30118588 Free PMC article.
References
-
- Romeo, V (2012). Myotonic Dystrophy Type 1 or Steinert's disease. Adv Exp Med Biol 724: 239–257. - PubMed
-
- Brook, JD, McCurrach, ME, Harley, HG, Buckler, AJ, Church, D, Aburatani, H et al. (1992). Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3' end of a transcript encoding a protein kinase family member. Cell 69: 385. - PubMed
-
- Mahadevan, M, Tsilfidis, C, Sabourin, L, Shutler, G, Amemiya, C, Jansen, G et al. (1992). Myotonic dystrophy mutation: an unstable CTG repeat in the 3' untranslated region of the gene. Science 255: 1253–1255. - PubMed
-
- Fu, YH, Pizzuti, A, Fenwick, RG Jr, King, J, Rajnarayan, S, Dunne, PW et al. (1992). An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science 255: 1256–1258. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

