Structural Analysis of dsRNA Binding to Anti-viral Pattern Recognition Receptors LGP2 and MDA5
- PMID: 27203181
- PMCID: PMC4885022
- DOI: 10.1016/j.molcel.2016.04.021
Structural Analysis of dsRNA Binding to Anti-viral Pattern Recognition Receptors LGP2 and MDA5
Abstract
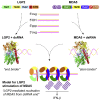
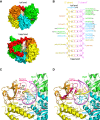
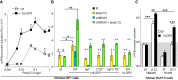
RIG-I and MDA5 sense virus-derived short 5'ppp blunt-ended or long dsRNA, respectively, causing interferon production. Non-signaling LGP2 appears to positively and negatively regulate MDA5 and RIG-I signaling, respectively. Co-crystal structures of chicken (ch) LGP2 with dsRNA display a fully or semi-closed conformation depending on the presence or absence of nucleotide. LGP2 caps blunt, 3' or 5' overhang dsRNA ends with 1 bp longer overall footprint than RIG-I. Structures of 1:1 and 2:1 complexes of chMDA5 with short dsRNA reveal head-to-head packing rather than the polar head-to-tail orientation described for long filaments. chLGP2 and chMDA5 make filaments with a similar axial repeat, although less co-operatively for chLGP2. Overall, LGP2 resembles a chimera combining a MDA5-like helicase domain and RIG-I like CTD supporting both stem and end binding. Functionally, RNA binding is required for LGP2-mediated enhancement of MDA5 activation. We propose that LGP2 end-binding may promote nucleation of MDA5 oligomerization on dsRNA.
Copyright © 2016 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures





Similar articles
-
Structural basis of double-stranded RNA recognition by the RIG-I like receptor MDA5.Arch Biochem Biophys. 2009 Aug 1;488(1):23-33. doi: 10.1016/j.abb.2009.06.008. Epub 2009 Jun 14. Arch Biochem Biophys. 2009. PMID: 19531363
-
Unraveling blunt-end RNA binding and ATPase-driven translocation activities of the RIG-I family helicase LGP2.Nucleic Acids Res. 2024 Jan 11;52(1):355-369. doi: 10.1093/nar/gkad1106. Nucleic Acids Res. 2024. PMID: 38015453 Free PMC article.
-
Solution structures of cytosolic RNA sensor MDA5 and LGP2 C-terminal domains: identification of the RNA recognition loop in RIG-I-like receptors.J Biol Chem. 2009 Jun 26;284(26):17465-74. doi: 10.1074/jbc.M109.007179. Epub 2009 Apr 20. J Biol Chem. 2009. PMID: 19380577 Free PMC article.
-
LGP2 synergy with MDA5 in RLR-mediated RNA recognition and antiviral signaling.Cytokine. 2015 Aug;74(2):198-206. doi: 10.1016/j.cyto.2015.02.010. Epub 2015 Mar 18. Cytokine. 2015. PMID: 25794939 Free PMC article. Review.
-
Structures of RIG-I-Like Receptors and Insights into Viral RNA Sensing.Adv Exp Med Biol. 2019;1172:157-188. doi: 10.1007/978-981-13-9367-9_8. Adv Exp Med Biol. 2019. PMID: 31628656 Review.
Cited by
-
Distinct and Orchestrated Functions of RNA Sensors in Innate Immunity.Immunity. 2020 Jul 14;53(1):26-42. doi: 10.1016/j.immuni.2020.03.017. Immunity. 2020. PMID: 32668226 Free PMC article. Review.
-
Cryo-EM Structures of MDA5-dsRNA Filaments at Different Stages of ATP Hydrolysis.Mol Cell. 2018 Dec 20;72(6):999-1012.e6. doi: 10.1016/j.molcel.2018.10.012. Epub 2018 Nov 15. Mol Cell. 2018. PMID: 30449722 Free PMC article.
-
Liquid-Liquid Phase Separation by Intrinsically Disordered Protein Regions of Viruses: Roles in Viral Life Cycle and Control of Virus-Host Interactions.Int J Mol Sci. 2020 Nov 28;21(23):9045. doi: 10.3390/ijms21239045. Int J Mol Sci. 2020. PMID: 33260713 Free PMC article. Review.
-
Uncovering targets of the Leader protease: Linking RNA-mediated pathways and antiviral defense.Wiley Interdiscip Rev RNA. 2021 Jul;12(4):e1645. doi: 10.1002/wrna.1645. Epub 2021 Feb 18. Wiley Interdiscip Rev RNA. 2021. PMID: 33605051 Free PMC article. Review.
-
Dynamic Evolution of Avian RNA Virus Sensors: Repeated Loss of RIG-I and RIPLET.Viruses. 2022 Dec 20;15(1):3. doi: 10.3390/v15010003. Viruses. 2022. PMID: 36680044 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

