Evolution of a transcriptional regulator from a transmembrane nucleoporin
- PMID: 27198230
- PMCID: PMC4888837
- DOI: 10.1101/gad.280941.116
Evolution of a transcriptional regulator from a transmembrane nucleoporin
Erratum in
-
Corrigendum: Evolution of a transcriptional regulator from a transmembrane nucleoporin.Genes Dev. 2017 Apr 15;31(8):845. doi: 10.1101/gad.300699.117. Genes Dev. 2017. PMID: 28512238 Free PMC article. No abstract available.
Abstract
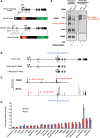
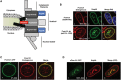
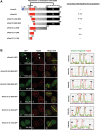
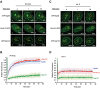
Nuclear pore complexes (NPCs) emerged as nuclear transport channels in eukaryotic cells ∼1.5 billion years ago. While the primary role of NPCs is to regulate nucleo-cytoplasmic transport, recent research suggests that certain NPC proteins have additionally acquired the role of affecting gene expression at the nuclear periphery and in the nucleoplasm in metazoans. Here we identify a widely expressed variant of the transmembrane nucleoporin (Nup) Pom121 (named sPom121, for "soluble Pom121") that arose by genomic rearrangement before the divergence of hominoids. sPom121 lacks the nuclear membrane-anchoring domain and thus does not localize to the NPC. Instead, sPom121 colocalizes and interacts with nucleoplasmic Nup98, a previously identified transcriptional regulator, at gene promoters to control transcription of its target genes in human cells. Interestingly, sPom121 transcripts appear independently in several mammalian species, suggesting convergent innovation of Nup-mediated transcription regulation during mammalian evolution. Our findings implicate alternate transcription initiation as a mechanism to increase the functional diversity of NPC components.
Keywords: Nup98; Pom121; evolution; hominoid; nuclear pore complex (NPC); transcription.
© 2016 Franks et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

Similar articles
-
NLS-mediated NPC functions of the nucleoporin Pom121.FEBS Lett. 2010 Aug 4;584(15):3292-8. doi: 10.1016/j.febslet.2010.07.008. Epub 2010 Jul 14. FEBS Lett. 2010. PMID: 20624389
-
Nuclear pore complex assembly and maintenance in POM121- and gp210-deficient cells.J Cell Biol. 2006 May 22;173(4):477-83. doi: 10.1083/jcb.200601002. Epub 2006 May 15. J Cell Biol. 2006. PMID: 16702234 Free PMC article.
-
Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm.Cell. 2010 Feb 5;140(3):360-71. doi: 10.1016/j.cell.2010.01.011. Cell. 2010. PMID: 20144760
-
Nucleoplasmic Nup98 controls gene expression by regulating a DExH/D-box protein.Nucleus. 2018 Jan 1;9(1):1-8. doi: 10.1080/19491034.2017.1364826. Epub 2017 Sep 21. Nucleus. 2018. PMID: 28934014 Free PMC article. Review.
-
The plant nuclear envelope and regulation of gene expression.J Exp Bot. 2015 Mar;66(6):1673-85. doi: 10.1093/jxb/erv023. Epub 2015 Feb 13. J Exp Bot. 2015. PMID: 25680795 Review.
Cited by
-
Nuclear Periphery Takes Center Stage: The Role of Nuclear Pore Complexes in Cell Identity and Aging.Neuron. 2020 Jun 17;106(6):899-911. doi: 10.1016/j.neuron.2020.05.031. Neuron. 2020. PMID: 32553207 Free PMC article. Review.
-
In Pursuit of Distinctiveness: Transmembrane Nucleoporins and Their Disease Associations.Front Oncol. 2021 Dec 14;11:784319. doi: 10.3389/fonc.2021.784319. eCollection 2021. Front Oncol. 2021. PMID: 34970494 Free PMC article. Review.
-
Advances in the understanding of nuclear pore complexes in human diseases.J Cancer Res Clin Oncol. 2024 Jul 30;150(7):374. doi: 10.1007/s00432-024-05881-5. J Cancer Res Clin Oncol. 2024. PMID: 39080077 Free PMC article. Review.
-
Nuclear Pores Promote Lethal Prostate Cancer by Increasing POM121-Driven E2F1, MYC, and AR Nuclear Import.Cell. 2018 Aug 23;174(5):1200-1215.e20. doi: 10.1016/j.cell.2018.07.015. Epub 2018 Aug 9. Cell. 2018. PMID: 30100187 Free PMC article.
-
Nuclear pore complexes - a doorway to neural injury in neurodegeneration.Nat Rev Neurol. 2022 Jun;18(6):348-362. doi: 10.1038/s41582-022-00653-6. Epub 2022 Apr 29. Nat Rev Neurol. 2022. PMID: 35488039 Free PMC article. Review.
References
-
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. 2007. The molecular architecture of the nuclear pore complex. Nature 450: 695–701. - PubMed
-
- Antonin W, Franz C, Haselmann U, Antony C, Mattaj IW. 2005. The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol Cell 17: 83–92. - PubMed
-
- Brawand D, Soumillon M, Necsulea A, Julien P, Csardi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, et al. 2011. The evolution of gene expression levels in mammalian organs. Nature 478: 343–348. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
